
Tutorial
I. Obtaining RIscoper
II. Launching RIscoper
III. Quick start with example file
IV. Input
V. Parameter setting
VI. RIscoper execution and presentation
VII. Sentence and RNA name upload
VIII. Running RIscoper from web server
IX. Running RIscoper from the Command Line (Linux, Unix, DOS)
USER GUIDE
FAQ
I. Obtaining RIscoper
Instructions for obtaining RIscoper are available on the download page. In addition, the web server for RIscoper is freely available.
II. Launching RIscoper
To begin using RIscoper, follow the instructions on the download page for ensuring that you have Java on your computer and for downloading RIscoper. It is recommended that your computer has Java version 1.8 or later and your computer has at least 2 gigabytes of RAM.
When RIscoper starts, the main window appears. The main components of the user interface are as shown below:
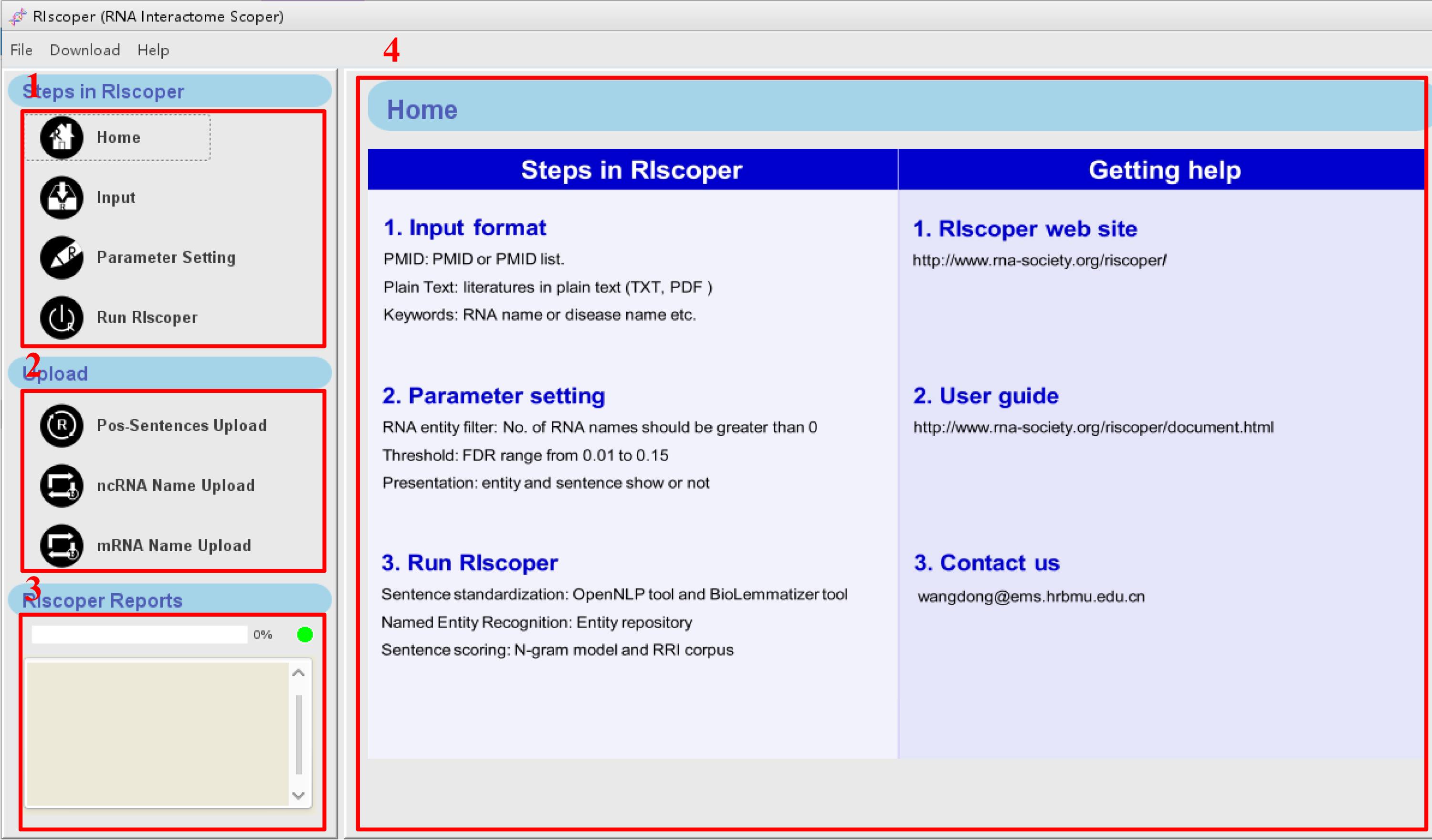
1. The navigation bar, which provides quick access to common RIscoper operations.
2. The upload bar, which is used to upload new RNA names and positive RRI sentences to the system.
See Sentence and RNA name upload for
details .
3. The processes panel, which provides information about the status of your analyses.
4. The main panel, which is used to display dialogs and results. When you start RIscoper, the main panel displays the Home page.
III. Quick start with example files
Once you have downloaded and started RIscoper,
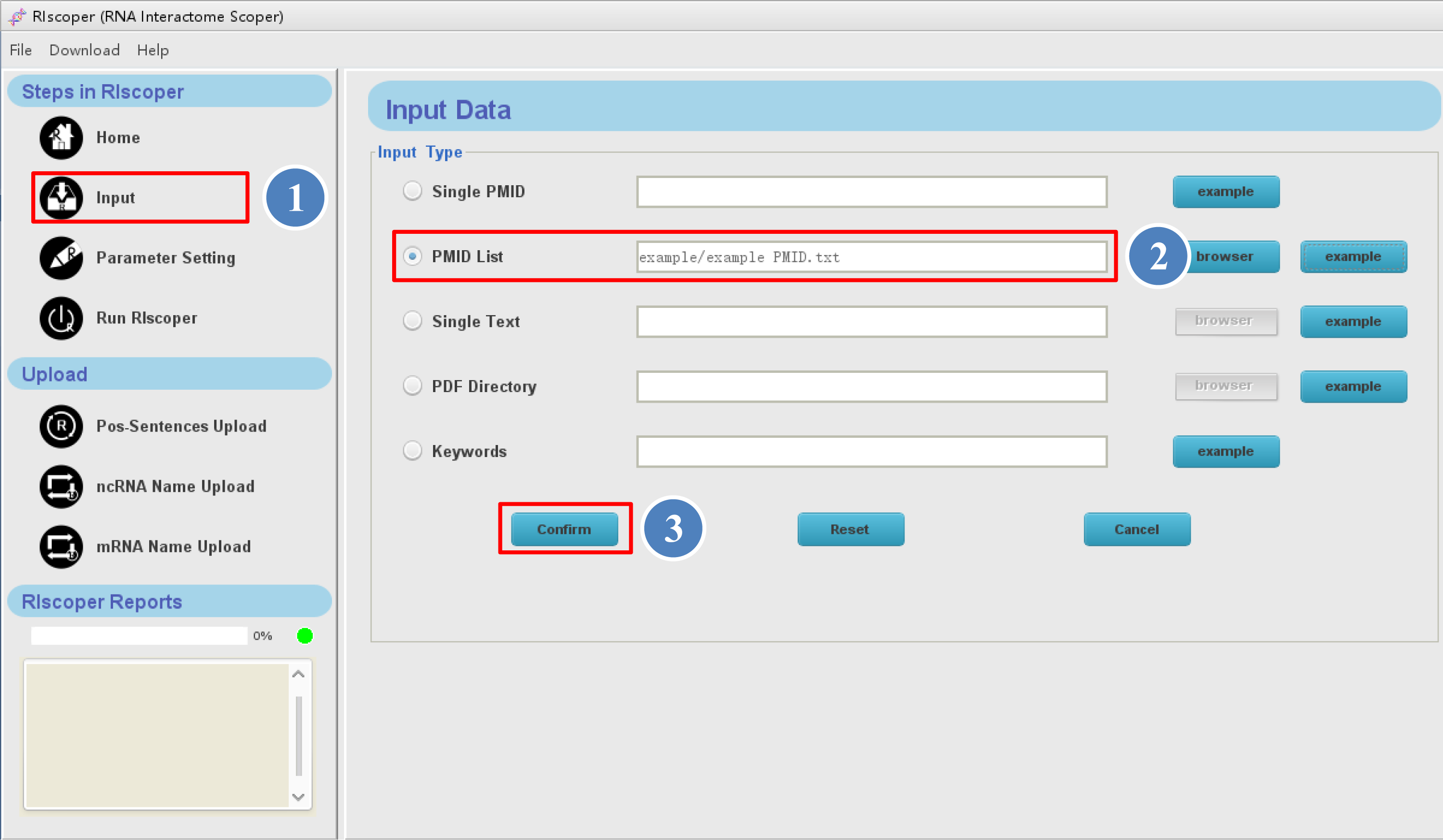
① click "Input" icon in the navigation
bar, you will see the Input interface as shown below, you can input the literatures in different ways (see Input for details). Here, we provide a PMID list (example PMIDs.txt) as an example,
② select the "PMID list" icon, and click "browser" button to upload the example PMIDs file from your computer (or you can just click "example" button instead),
③ then click "Confirm" button to download the article abstracts. It may take some time for RIscoper to automatically download the article abstracts from PubMed, depending on number of the articles and your internet speed.
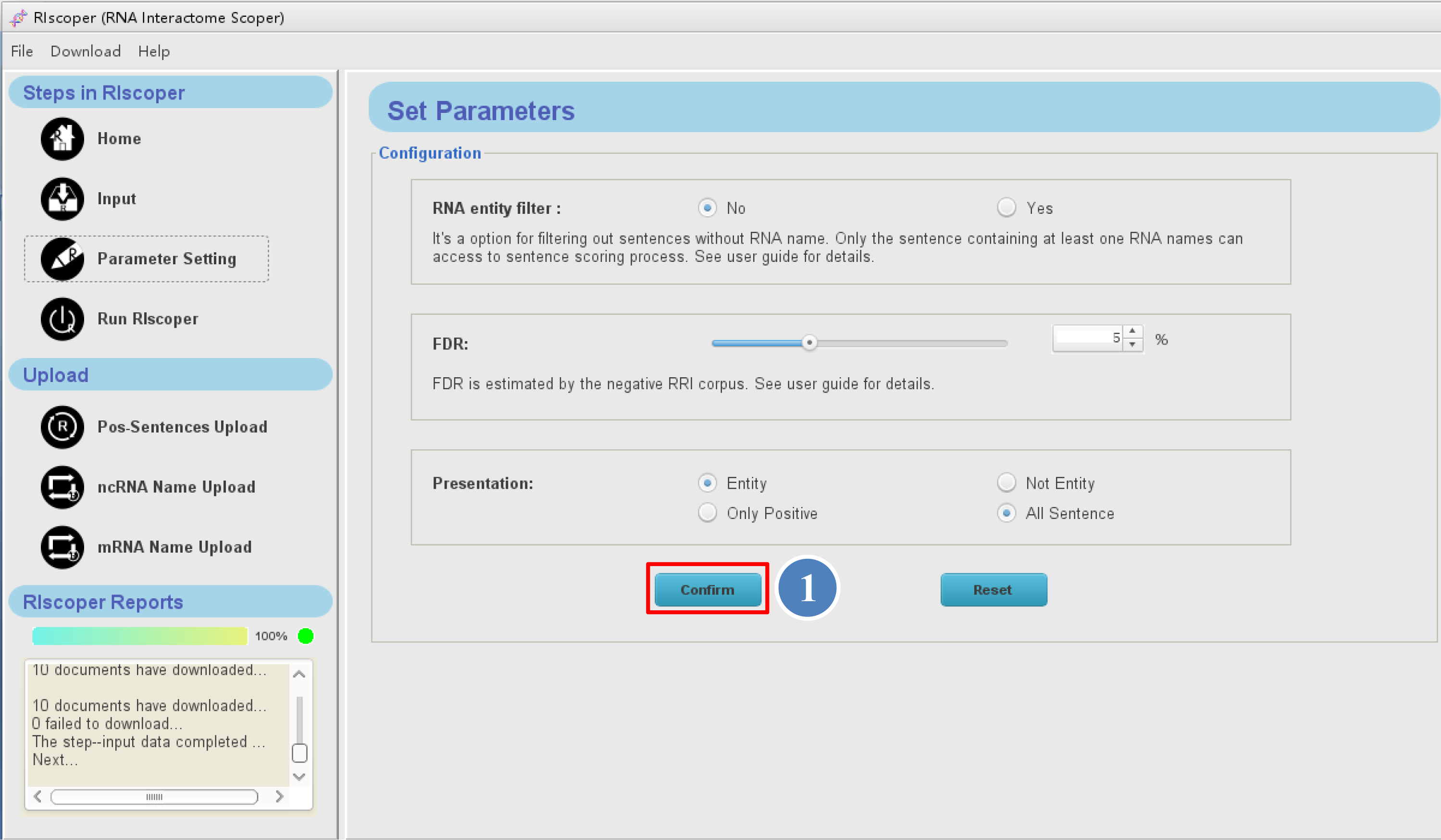
Once downloaded, the window will go to the Parameter setting interface as shown
below. You can set the parameters for your analysis on this interface (see Parameter setting for details). Here, we start the analysis with default parameters,
④ by click "Confirm" button, the window will go to the Run RIscoper interface.
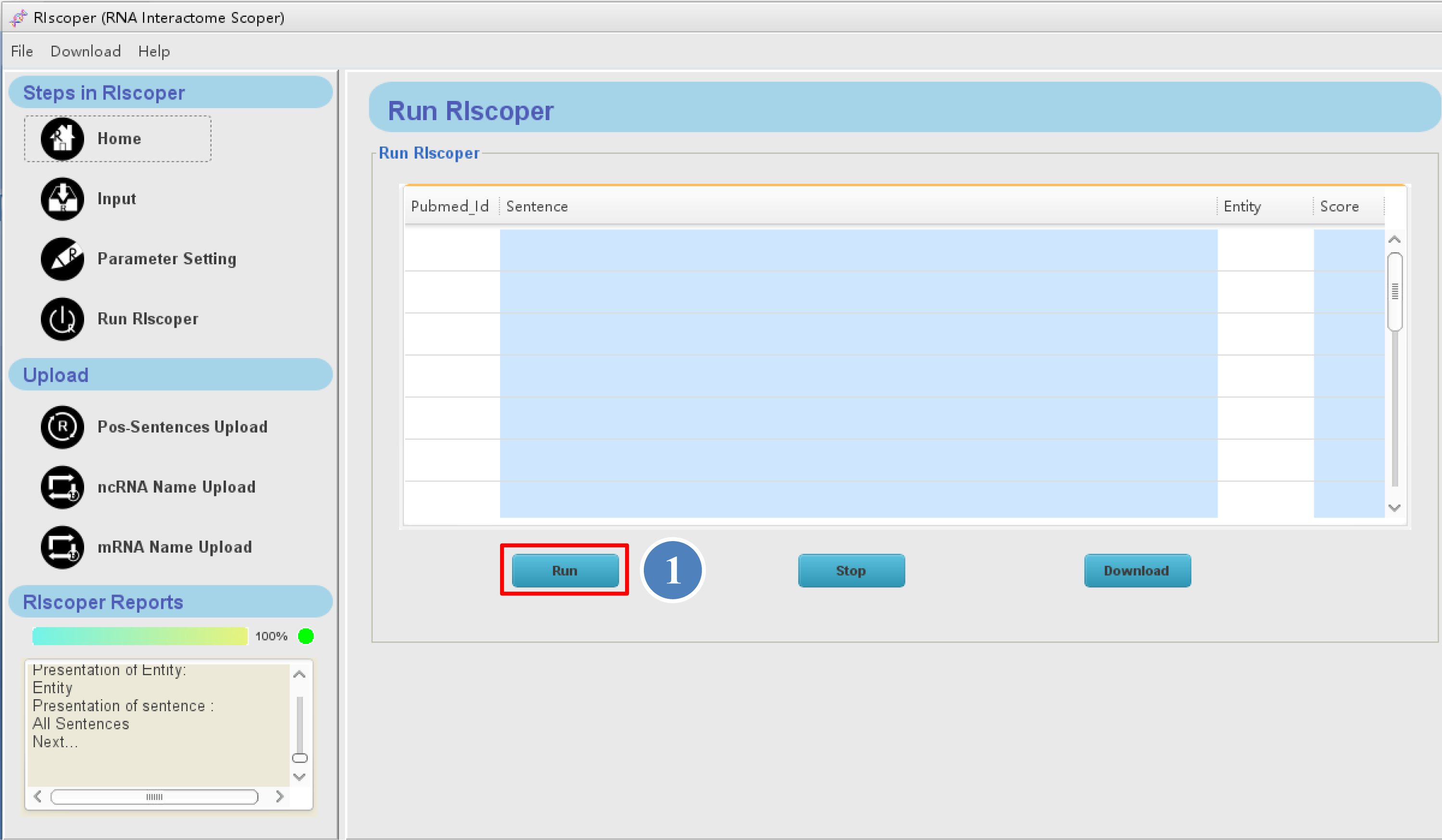
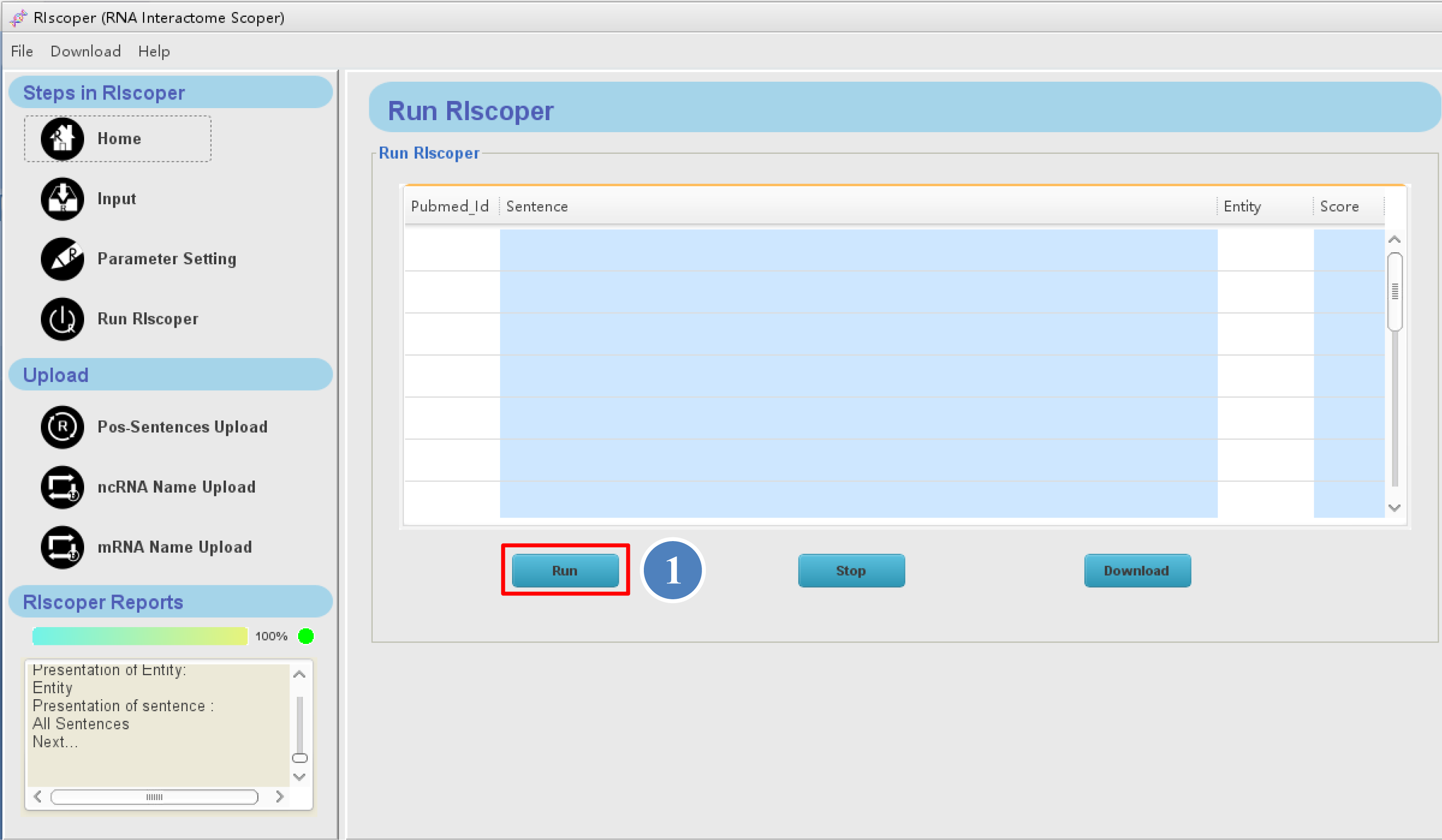
The Run RIscoper interface is as shown below,
⑤ you can click the "Run" button to
execute your analysis. It may take a few minutes for RIscoper to complete the analysis, depending on number of the articles.
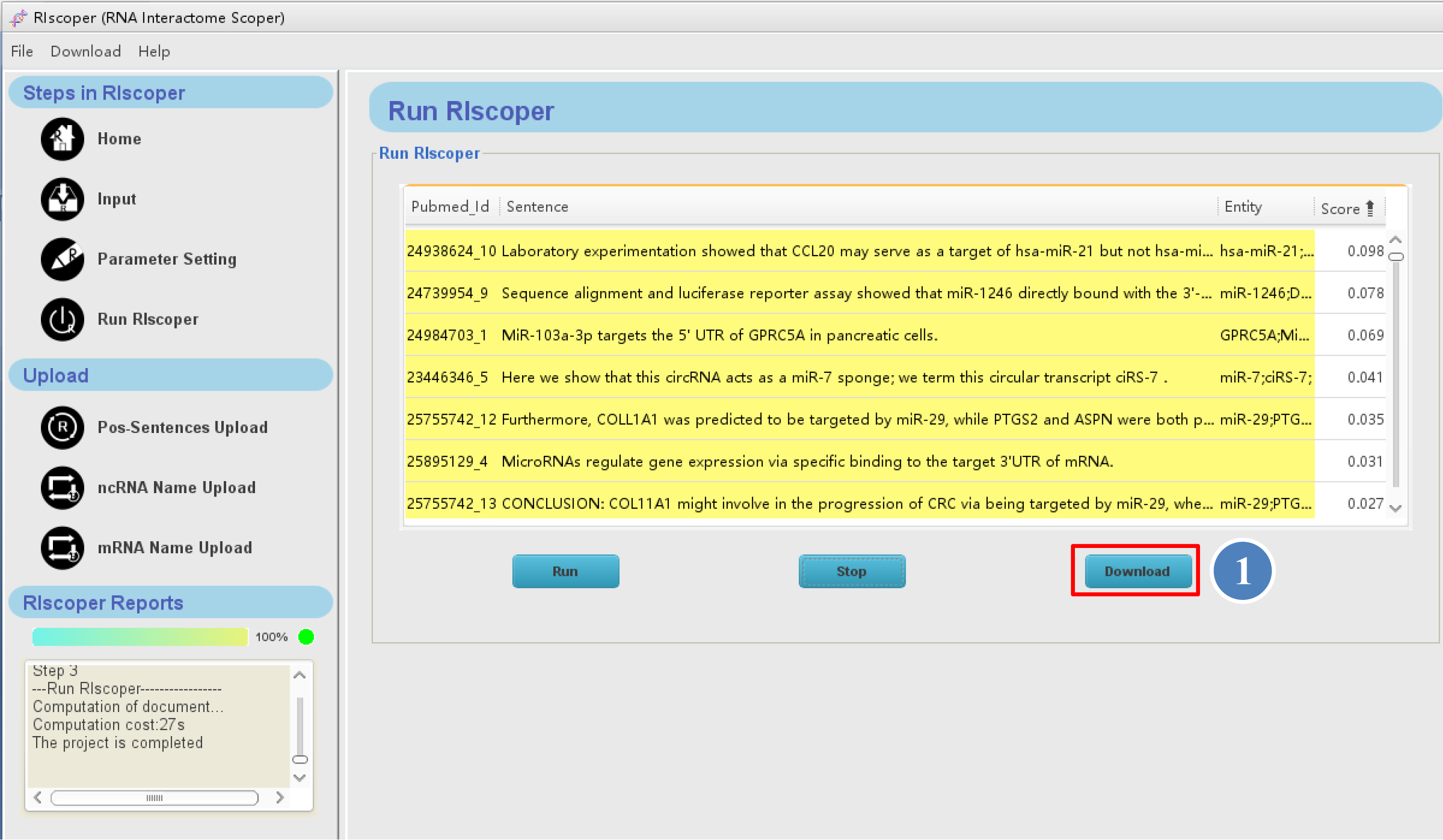
When the analysis is complete, the results will display in the main panel as shown below. The results table contains sentences inputted and corresponding PMIDs, scores and entity names (see RIscoper execution and output for details).
⑥ You can browse the result on RIscoper or download it to your computer by click "Download" button.
IV. Input
Click "Input" icon in the navigation bar, you will see the Input interface as shown below. RIscoper allows users to upload full texts or abstracts, and provides an online search tool that is connected with PubMed (PMID and keywords input), and these capabilities are useful for biologists. Furthermore, RIscoper provides "example" button for each input way.
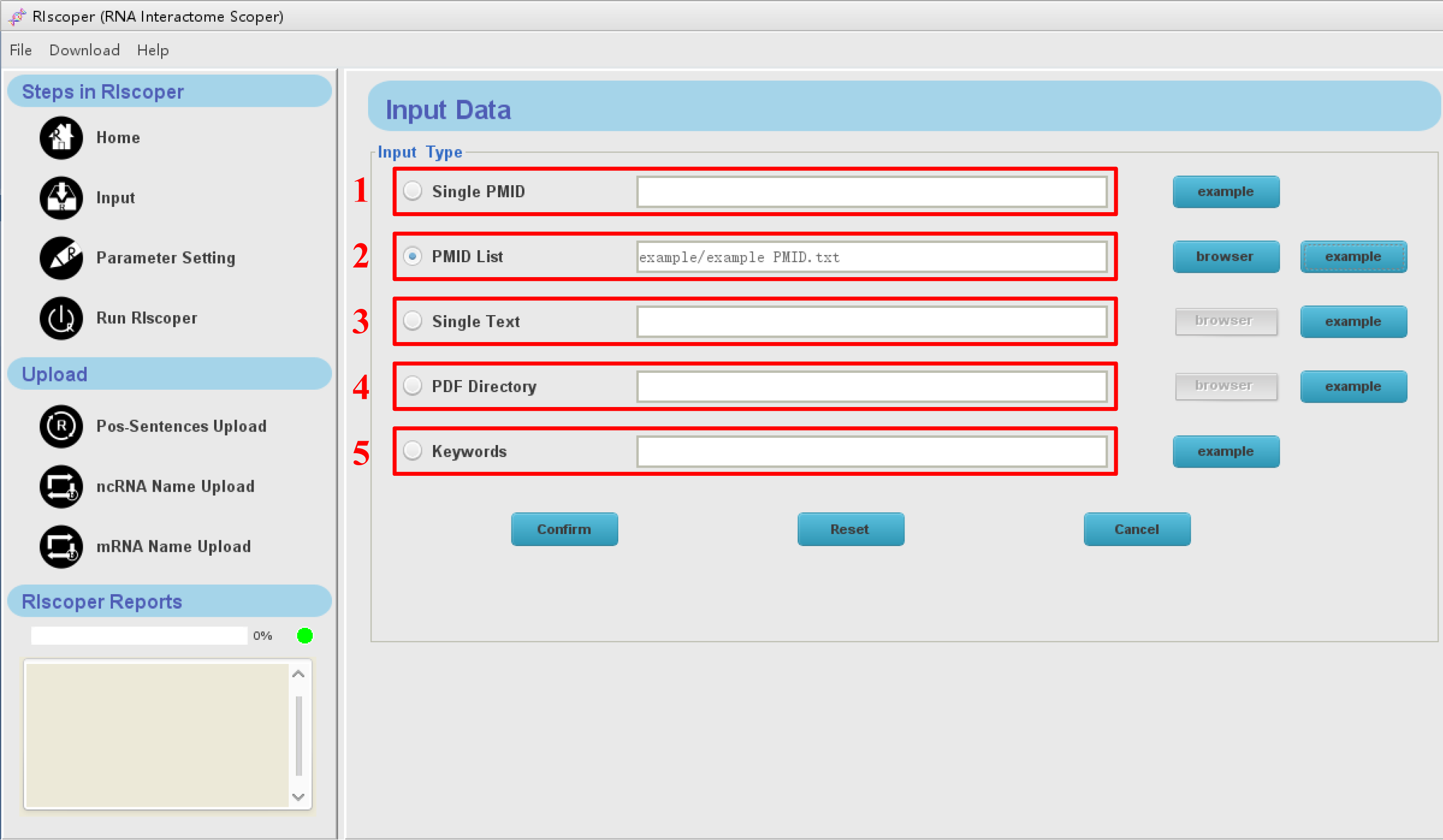
1. Single PMID: a single PMID can be fill in the box (e.g: PMID: 24984703).
2. PMID list: list of PMIDs in plain text can be submitted (e.g: example PMID.txt).
3. Single text: literatures in plain text can be submitted (e.g: example articles.txt).
4. PDF directory: a folder of PDF documents can be submitted (e.g: example PDF file.zip).
5. Keywords: a keyword can be fill in the box (e.g: mir-34a-5p).
V. Parameter setting
Click "Parameter setting" icon in the navigation bar, you will see the Parameter setting interface as shown below. There are three parameters for configuration.
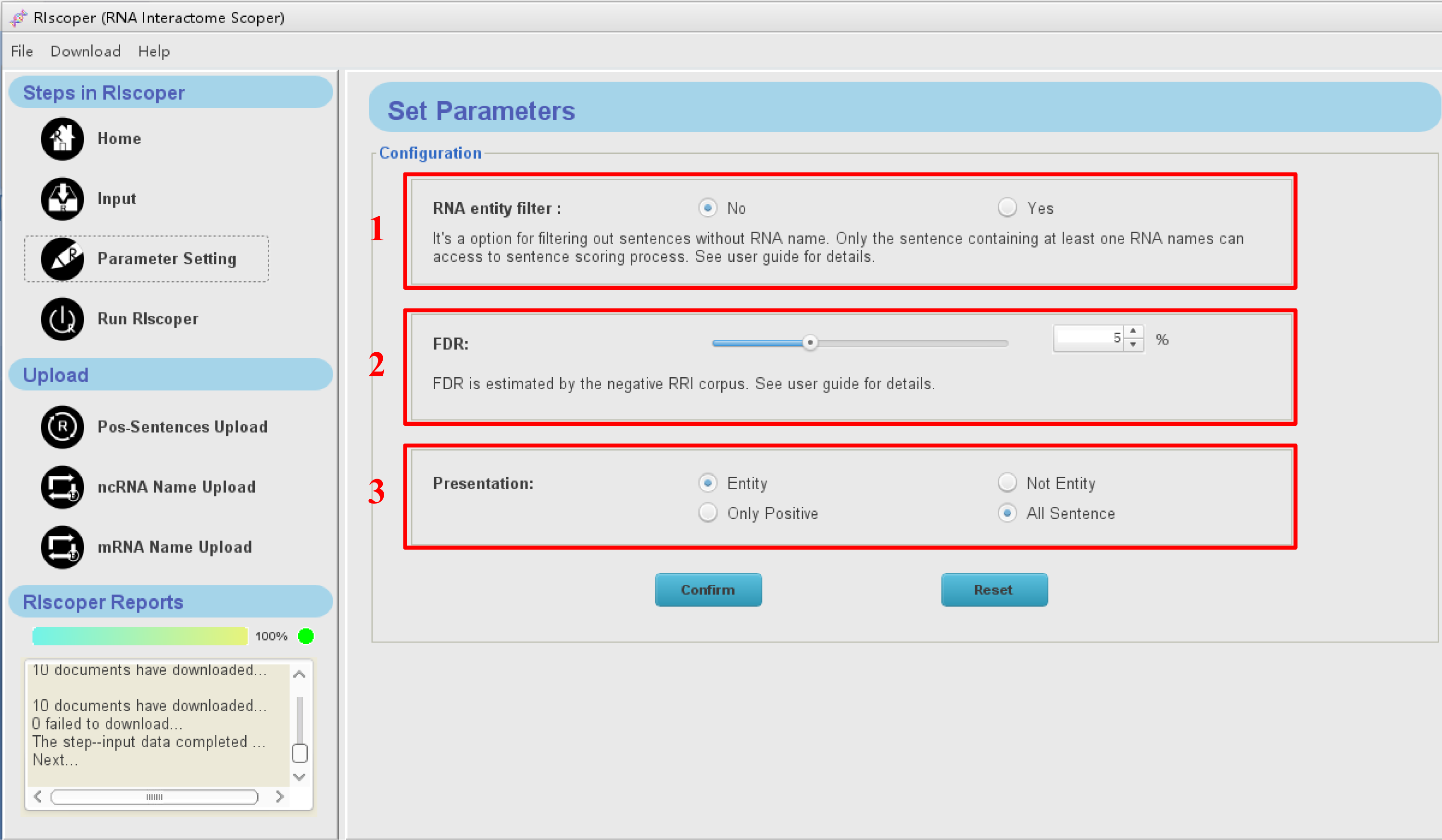
1. RNA entity filter:
- It’s a option for filtering out sentences without RNA name. Only the sentence containing at least one RNA names can access to sentence scoring process.
- In most cases, the RNA entity filter could improve the performance and speed of RIscoper. However, due to the limited coverage of the preset entity repository, some positive sentences may be mistaken deleted because of contain RNA names (new or unconventional name usually) have not been included in the preset entity repository. So selection of this filter or not need a trade off based on user requirements.
2. FDR:
- The FDR threshold is estimated by the negative RRI corpus, range from 1% to 15%. For users focused on one or several RNAs, the preferred FDR threshold value is 5%. For users seeking to extract as many RRIs as possible from a large number of studies, the preferred FDR threshold value is 10% or larger.
3. Presentation:
- Entity show: it allow users to select shown the entity names from sentence or not.
- Sentence show: show all the sentences inputted or only the sentences pass the FDR Threshold.
VI. RIscoper execution and presentation
It’s so easy to run RIscoper.
With click "Run" button in the navigation bar, the analysis begin to execution, then you can see the status of analysis by the processes panel in the bottom left corner. The calculating speed of RIscoper has been tested on a PC with a 2.8 GHz CPU and 8 GB RAM, the results demonstrated that RIscoper can run on PC with fast calculating speed (it tooks about 23s for 100 sentences, 70s for 1000 sentences, 105s for 5000 sentences and 148s for 10000 sentence).
The details of computational procedure consisting of three steps:
- (a). Sentence standardization, the articles inputted will be segmented into sentences by OpenNLP tool and all words will be lemmatized by BioLemmatizer tool. Then RIscoper deletes all words in brackets, strips whitespace and punctuation except commas and periods, and converts all words to lowercase.
- (b). Named Entity Recognition, a preset entity repository (collected a large number of RNA names from various databases and corpuses) is employed to recognize the RNA names in each sentence.
- (c). Sentence scoring, the sentence is scored by N-gram statistical model and a manually curated RRI corpus (containing 13377 sentences with RRI information). And then, Katz smooth and geometric mean algorithm are used to smoothed and normalized the score, respectively.
When the analysis is complete, RIscoper display the results by table format as shown below.
① You can browse the result on RIscoper or download it to your computer by click "Download"
button.
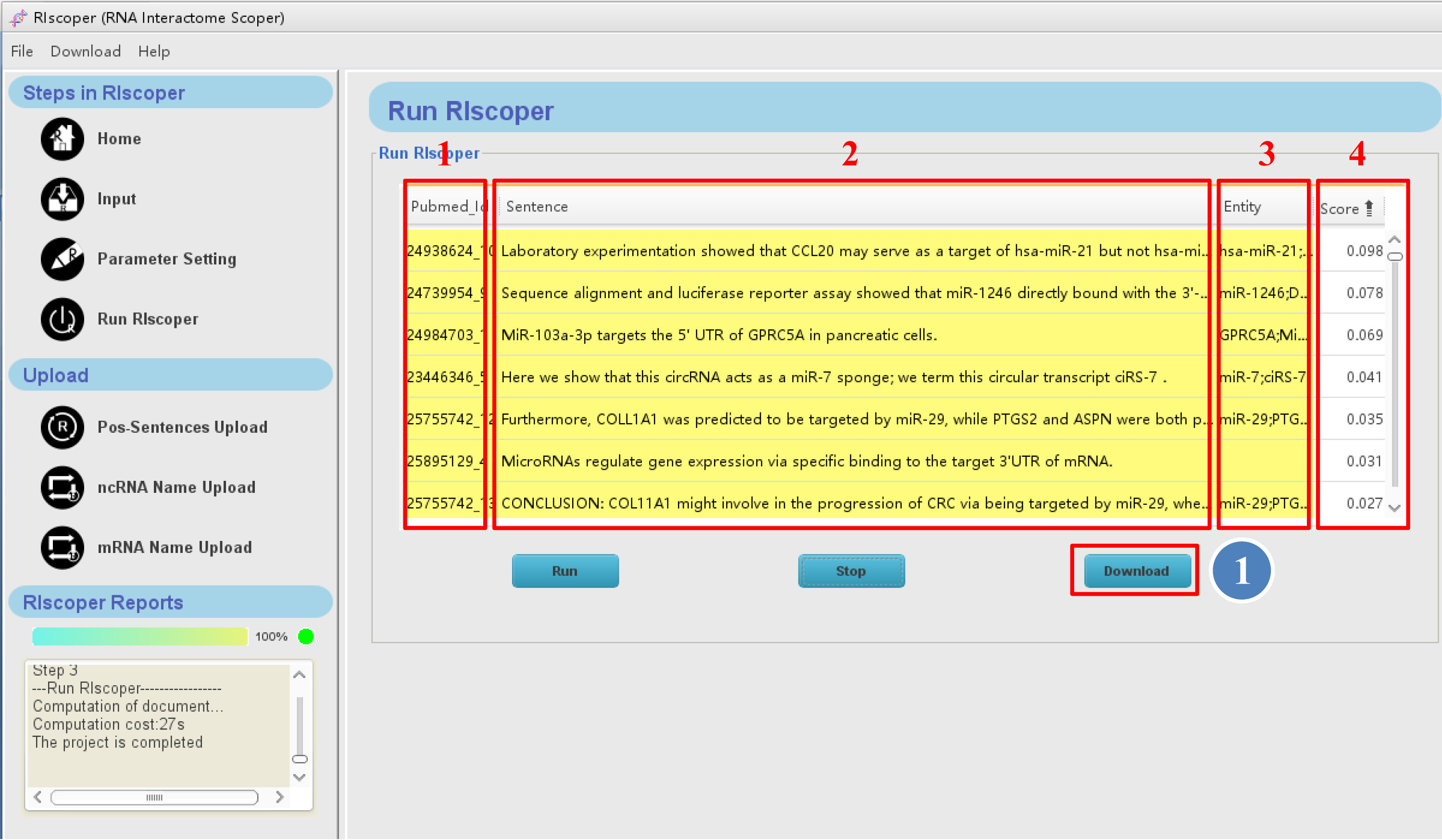
1. The first column of the table contains PMID and the numerical order of sentences (e.g, 24635082_1 means the sentence is the first sentence in the article with PMID 24635082).
2. The second column of the table contains analyzed sentences.
3. The third column of the table contains entity names (RNA names) of sentences.
4. The fourth column of the table contains score of sentences, which ranges from 0 to 1, with a higher score indicating more possibility of containing RRI information.
VII. Sentence and RNA name upload
RIscoper provide options for uploading new positive RRI sentences and RNA names to extend the
RRI corpus and/or entity repository. You can upload your own RRI sentences and RNA names by the
upload buttons, then RIscoper can automated integration of them to pre-existing RRI corpus
and/or entity repository for your analysis.
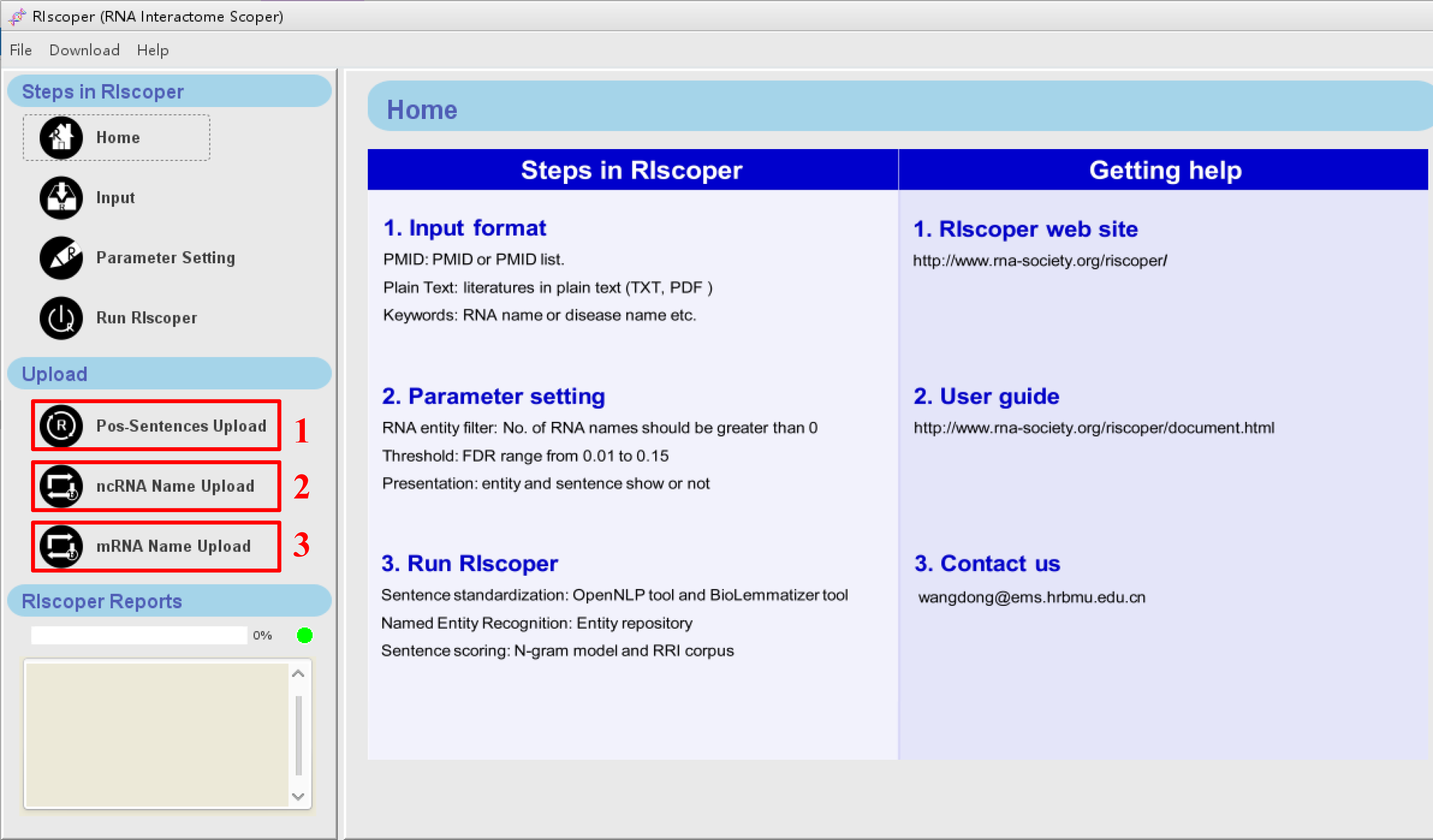
1. Pos-sentence upload: for uploading new RRI sentences (pos-sentence.txt).
2. ncRNA upload: for uploading new ncRNA names (ncRNA-entity.txt).
3. mRNA upload: for uploading new mRNA names (
mRNA-entity.txt).
VIII. Running RIscoper from web server
The options of web server are as follows:
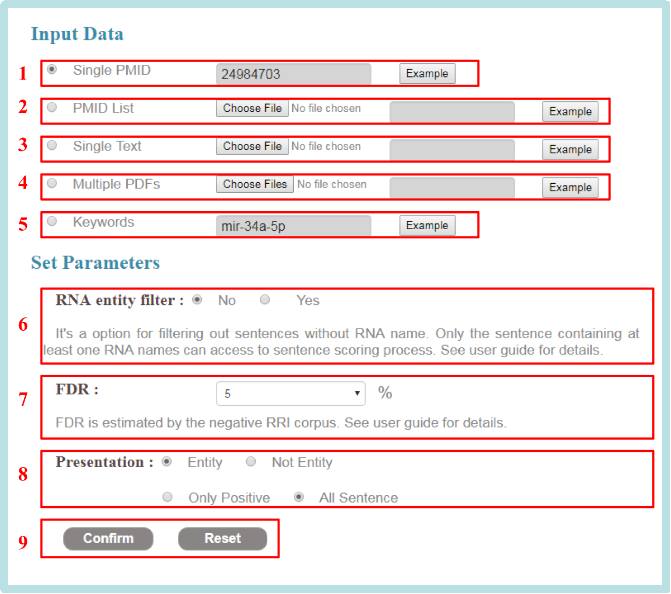
1. Single PMID: a single PMID can be fill in the box (e.g: PMID: 24984703).
2. PMID list: list of PMIDs in plain text can be submitted (e.g:example PMID.txt).
3. Single text: literatures in plain text can be submitted (e.g:example articles.txt).
4.Multiple PDFs: Multiple PDF documents can be submitted (e.g:example PDF file.zip).
5. Keywords: a keyword can be fill in the box (e.g: mir-34a-5p).
6. RNA entity filter: It’s a option for filtering out sentences without RNA name. Only the sentence containing at least one RNA names can access to sentence scoring process.
7. FDR: The FDR threshold is estimated by the negative RRI corpus, range from 1% to 15%. For users focused on one or several RNAs, the preferred FDR threshold value is 5%. For users seeking to extract as many RRIs as possible from a large number of studies, the preferred FDR threshold value is 10% or larger.
8. Presentation: (1) Entity show: show the entity names in sentences or not. (2) Sentence show: show all the sentences inputted or only the sentences pass the FDR threshold.
9. Confirm and reset option, run the tool by click "confirm" or reset the tool by click "reset".
IX. Running RIscoper from the Command Line (Linux, Unix, DOS)
Typically, you run RIscoper software using the RIscoper desktop application; however, you can also run RIscoper software from the command line (for Linux, Unix, DOS).
Command form
To run RIscoper from the command line, use a java command of the form:
java -Xmx1G -Dfile.encoding=utf-8 -jar full path/RIscoper.jar -i <"full path/input_filename.txt"> -o "full path/output_filename.txt" -t <"i"|"il"|"t"|"p"|"k"> -f <"yes"|"no"> -p <"integer"> -s <"yes"|"no">
Syntax
- -i (--input)
- -o (--output)
- -t (--type)
- -f (--filter)
- -p (--fdr)
- -s (--sentence)
- -e (--entity)
- -u (--upload)
- -h (--help)
| | Input content file (PMID, articles, keywords etc.). |
| Usage: -i <"PMID"> | |
| -i <"keyword"> | |
| -i <"full path/input_filename.txt"> | |
| -i <"full path"> | |
| | Output the results to a text document (.txt). |
| Usage: -o <"full path/output_filename.txt"> | |
| | Select input types, there are five types as follows |
| i, input the PMID, for example "24984703"; | |
| il, input the PMID list, for example: " example PMID.txt "; | |
| t, input the articles file, for example: "example articles.txt"; | |
| p, input the PDF files, for example: "/example PDF"; | |
| k, input the keyword, for example: " mir-34a-5p "; | |
| Usage: -t <"i"|"il"|"t"|"p"|"k"> | |
| | Select the "RNA entity filter" or not: |
| yes: select "RNA entity filter"; | |
| no: not select "RNA entity filter", the default. | |
| Usage: -f <"yes"|"no"> | |
| | Set the FDR threshold (range from 0 to 15). |
| Usage: -p <"integer"> 0 ≤ integer ≤ 15 | |
| | Only export the positive sentences (sentence with score higher than FDR threshold) or export all sentences: |
| yes: only export the positive sentences; | |
| no: export all sentences. the default. | |
| Usage: -s <"yes"|"no"> | |
| | Export the sentences containing entities which are identified from sentences of input file or export the sentences without entities: |
| yes: export the sentences containing entities. default; | |
| no: export the sentences without entities. | |
| Usage: -e <"yes"|"no"> | |
| | Upload new positive RRI sentences and RNA names. |
| sentence, upload new positive RRI sentences. | |
| ncRNA, upload new ncRNA names. | |
| mRNA, upload new mRNA names. | |
| Usage: -u <"sentence"|"ncRNA"|"mRNA"> | |
| | Get help for all Syntax. |
| Usage: -h <"help"> |
Examples:
Input PMID (PMID: "24984703"):
java -Xmx1G -Dfile.encoding=utf-8 -jar full path/RIscoper.jar -i "24984703" -o "full path/output.txt" -t "i" -f "no" -p "5" -s "yes"
Input PMID list (example PMID.txt):
java -Xmx1G -Dfile.encoding=utf-8 -jar full path/RIscoper.jar -i "full path/example PMID.txt" -o "full path/output.txt" -t "il" -f "no" -p "5" -s "yes"
Input articles (example articles.txt):
java -Xmx1G -Dfile.encoding=utf-8 -jar full path/RIscoper.jar -i "full path/articles.txt" -o "full path/output.txt" -t "t" -f "no" -p "5" -s "yes"
Input PDF files ("example PDF" folder):
java -Xmx1G -Dfile.encoding=utf-8 -jar full path/RIscoper.jar -i "full path/example PDF" -o " full path/output.txt" -t "p" -f "no" -p "5" -s "yes"
Input keyword (keyword: "mir-34a-5p"):
java -Xmx1G -Dfile.encoding=utf-8 -jar full path/RIscoper.jar -i "mir-34a-5p" -o "full path/output.txt" -t "k" -f "no" -p "5" -s "yes"
Upload new sentences:
java -Xmx1G -Dfile.encoding=utf-8 -jar full path/RIscoper.jar -i "full path/uplaod_sentences.txt" -u "sentence"
Q: What is the purpose of RIscoper?
A: RIscoper is a user friendly software written in JAVA, which is a fast and simple tool that extracted RRIs from literatures with high performance and practicability.In addition, the web server for RIscoper is freely available.
Q: How do I use RIscoper?
A: For basic instructions on using RIscoper, see the User Guide.
Q: What is requirements of computer configuration for RIscoper
A: RIscoper can works on multiple platforms (GUI version for Windows (for 32 and 64 bit) and Mac system, command line version for Linux, Unix and DOS). It is recommended that your computer has Java version 1.8 or later and your computer has at least 2 gigabytes of RAM. Moreover, the software with JRE are provided for windows and Mac system as well, which can work on systems without JRE.
Q: What does that RIscoper take as input?
A: RIscoper allows users to upload full texts or abstracts, and provides an online search tool that is connected with PubMed (PMID and keyword input), and these capabilities are useful for biologists.
Q: Can I use RIscoper without internet connection?
A: If there is no internet connection, the PMID input and keywords input of RIscoper are infeasible. Because of they need connect to the PubMed to download articles. However, you can use RIscoper by input articles in plain text (TXT or PDF) or a folder of plain texts which on local computer.
Q: If the download speed of articles is too slow when I use the PMID input and keywords Input, What should I do?
A: You can directly download all the articles you need from PubMed by yourself, and save these documentation in plain text, then submit it using plain text input.
Q: What is the RNA entity filter?
A: It’s a option for filtering out sentences without RNA names. Only the sentence containing at least one RNA names can access to sentence scoring process. In most cases, the RNA entity filter could improve the performance and speed of RIscoper. However, due to the limited coverage of the preset entity repository, some positive sentences may be mistaken deleted because of contain RNA names (new or unconventional name usually) have not been included in the preset entity repository. So selection of this filter or not need a trade off based on user requirements.
Q: What is the FDR threshold in the RIscoper?
A: The FDR threshold is estimated by the negative RRI corpus (13377 negative sentences without RRI information ). For instance, the score at top 5% of the negative RRI corpus is defined as threshold score of FDR 5%, the score at top 1% of the negative RRI corpus is defined as threshold score of FDR 1%. For users focused on one or several RNAs, the preferred FDR threshold value is 5%. For users seeking to extract as many RRIs as possible from a large number of studies, the preferred FDR threshold value is 10% or larger.
Q: How can I upload the new RNA names or RRI sentences to RIscoper?
A: RIscoper support users upload new positive RRI sentences and RNA names to extend the RRI corpus and/or entity repository. You can upload your own RRI sentences and RNA names by the upload buttons, then RIscoper can automated integration of them to pre-existing RRI corpus and/or entity repository for your analysis.
Q: How can I download the RRI corpus?
A: All the sentences in RRI corpus were manually confirmed carefully. As a first dataset describing RNA-RNA interactions, this dataset providing a favourable resource for ongoing text mining studies of RNA interactions and will be a useful dataset in other future machine learning work related to natural language processing. The corpus could be download at download page.
Q: What are the differences between RAID v2.0 database and RIscoper?
A:The RAID v2.0 database contains more than 5,200,000 RRIs from various sources, including manually curated RRIs from the literature, computational prediction RRIs by multiple algorithms and experimentally validated RRIs from other databases. However, RIscoper is a text-mining tool for RRI extraction, and it can instantly scan the latest research from the literature. Compared with RIscoper, the RAID database is usually unable to cover all RRI research findings, especially the current research status of certain RNAs. For users seeking to identify the interactions of certain RNAs from the literature, RIscoper is preferred. For users seeking to present all experimentally validated and predicted interactions of certain RNAs, RAID v2.0 is preferred.
Q: I discovered a software bug in RIscoper. What should I do?
A: Please report the bug to us immediately by email, we can assure you that each bug report will be addressed and responded as soon as possible. Contact address: wangdong @ems.hrbmu.edu.cn.