
Tutorial
The new development of ViRBase
1. Expand data sources and coverage of organism.
2. Support new functions of partial and batch searches.
3. Provide two interactome tools: IntaRNA and PRIdictor.
4. Add the RNA editing/localization/modification.
5. Add drug information, ncRNA SNP and interaction network.
The homepage is displayed in the following Fig.1-1.
Fig.1-1:
1. Main functions of the database are provided in menu bar form (boxed in light blue).
2. Other databases contributed by our group.
3. Cite information.
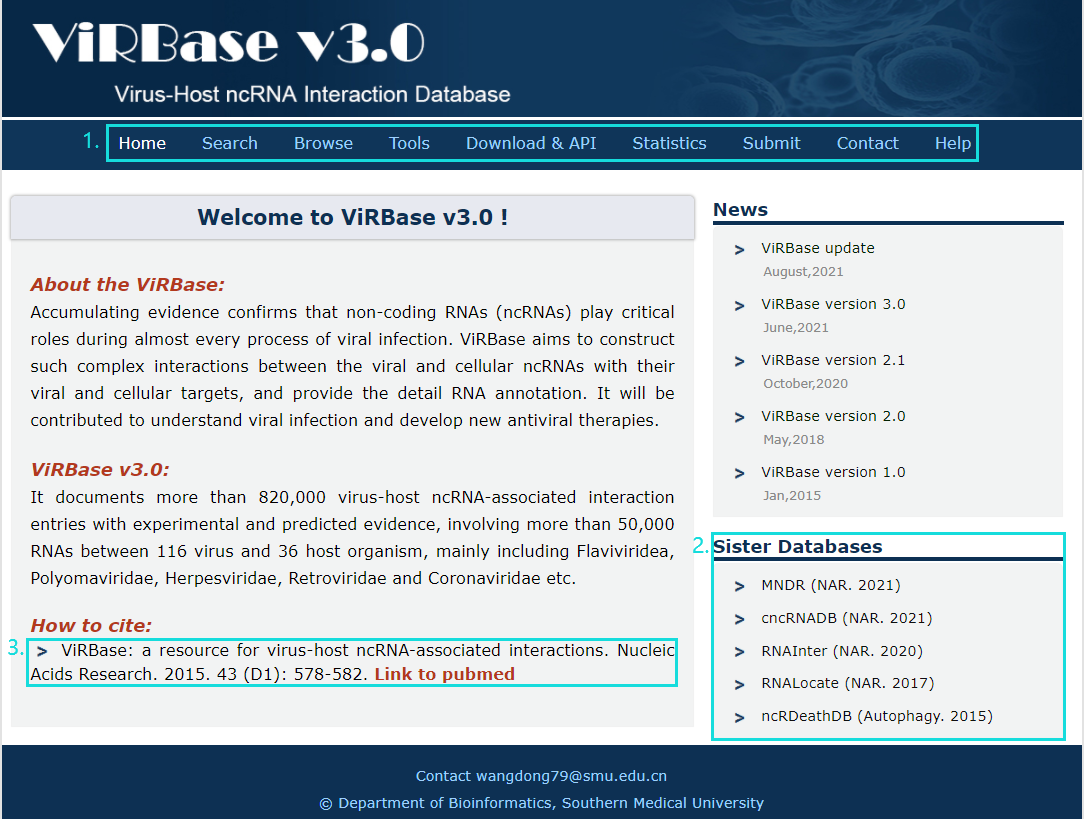
Fig.1-1 Homepage
The Exact search page is displayed in Fig.2-1:
1. Carefully select a dataset: Four choices are provided.
2. Enter a keyword corresponding to selected dataset.
3. Four categories provided to filter results: Interaction Type, Organism, Detection Method and Score.
4. Use NCBI Gene/ NCBI Taxonomy/miRBase to normalize your input information.
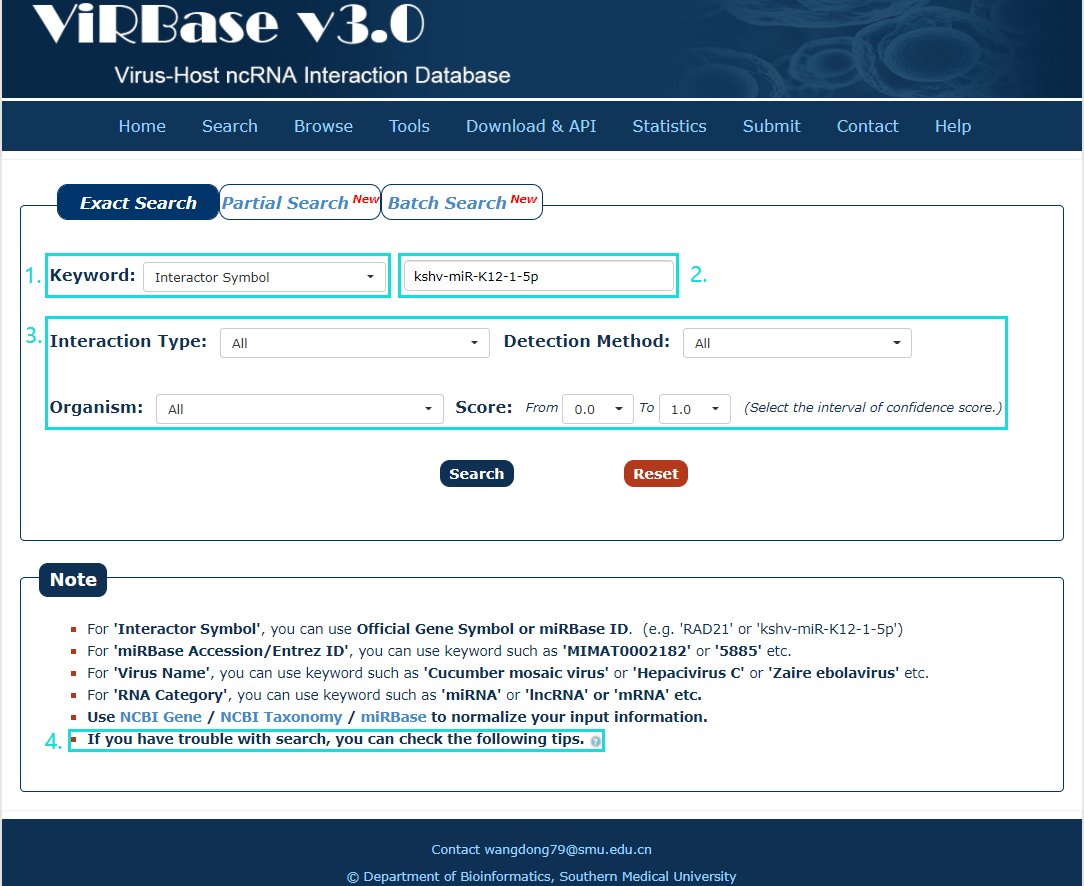
Fig.2-1 Exact Search page
The partial search page is displayed in Fig.2-2:
1. Select the category of your keyword.
2. Enter a keyword corresponding to select.
3. Choose the match entries what you want.
For example, we enter a keyword 'miR-7'.
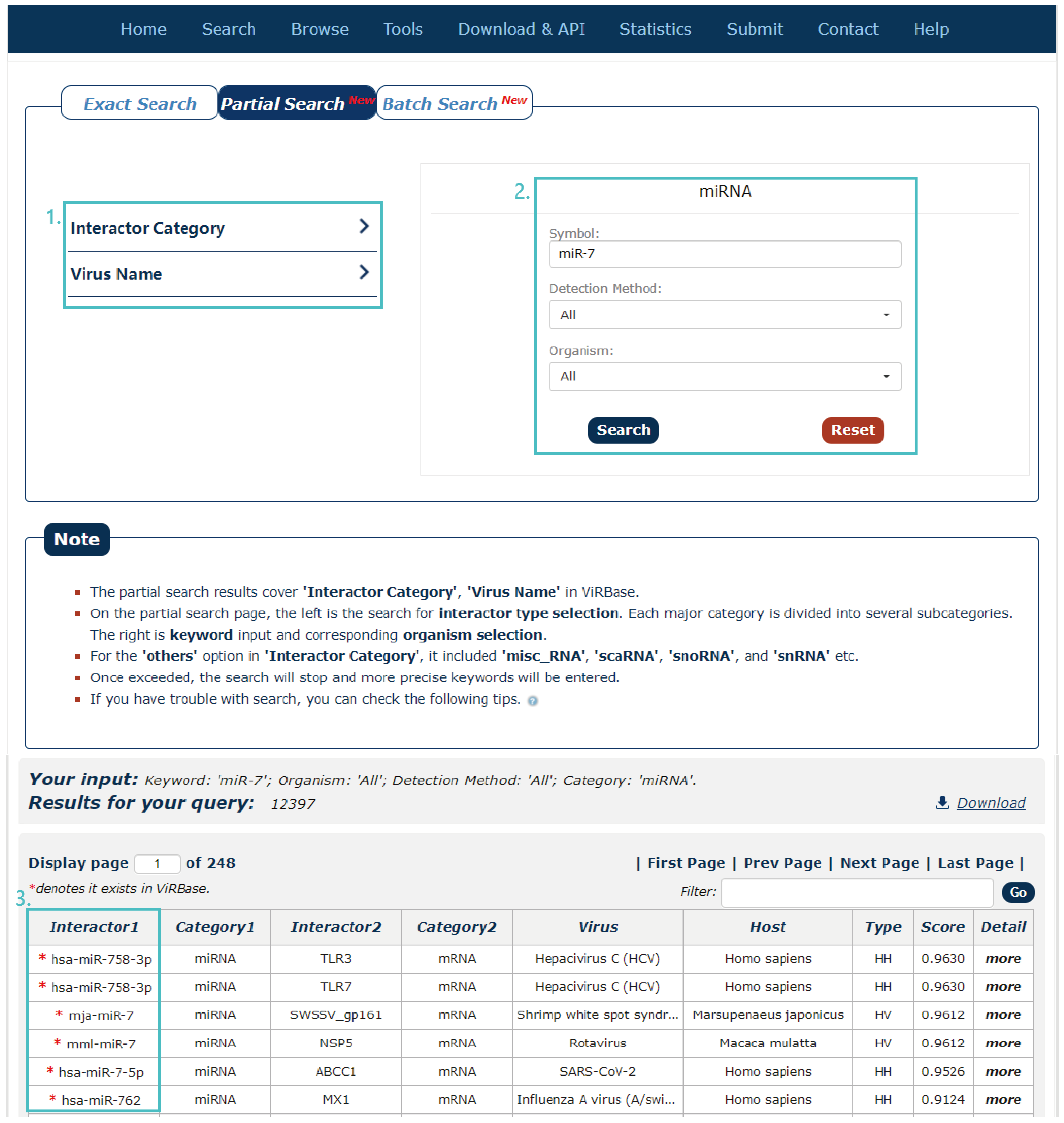
Fig.2-2 Partial Search page
The Batch search page is displayed in Fig.2-3:
1. Select a dataset for your keywords.
2. Enter the keywords or upload a file.
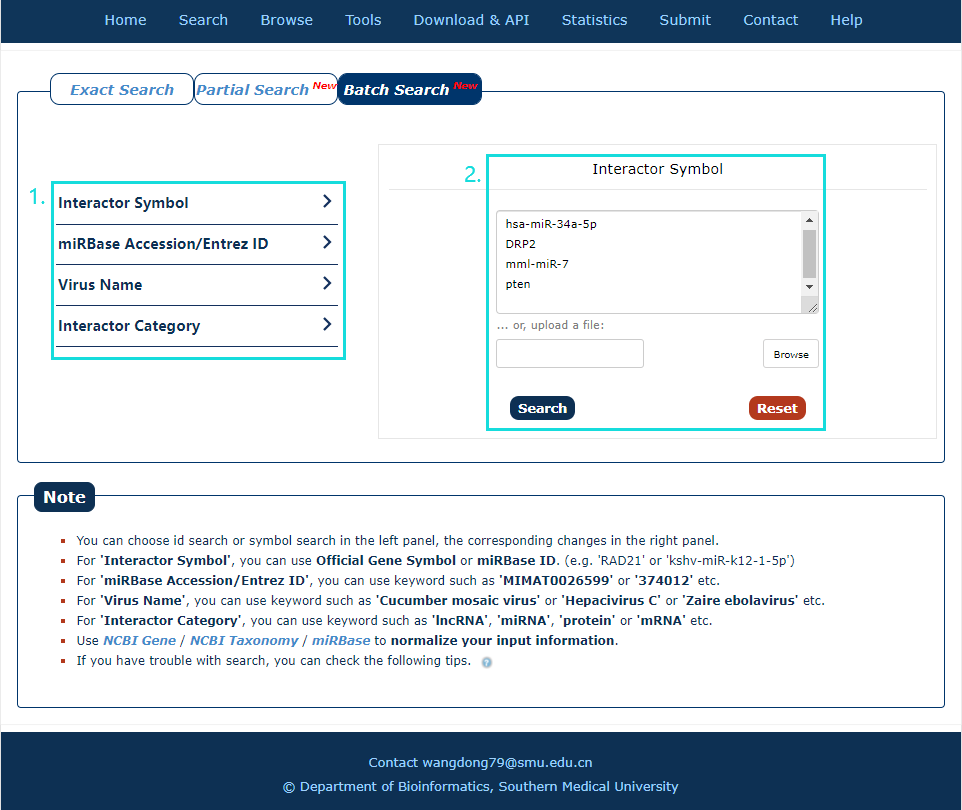
Fig.2-3 Batch Search page
This tutorial is as follows.
1. First we have to choose the type of keyword. There are three keyword types in our search as the picture shows. In this example, we choose Interactor Symbol as the keyword type.
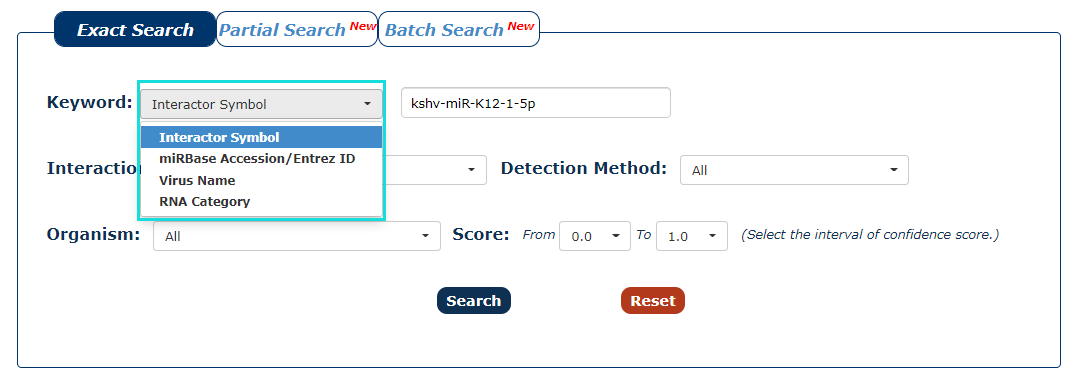
Fig.3-1
2. Next, we enter the keyword according to the keyword type selected in the previous step. In this example, we choose 'kshv-miR-K12-1-5p' as the keyword.
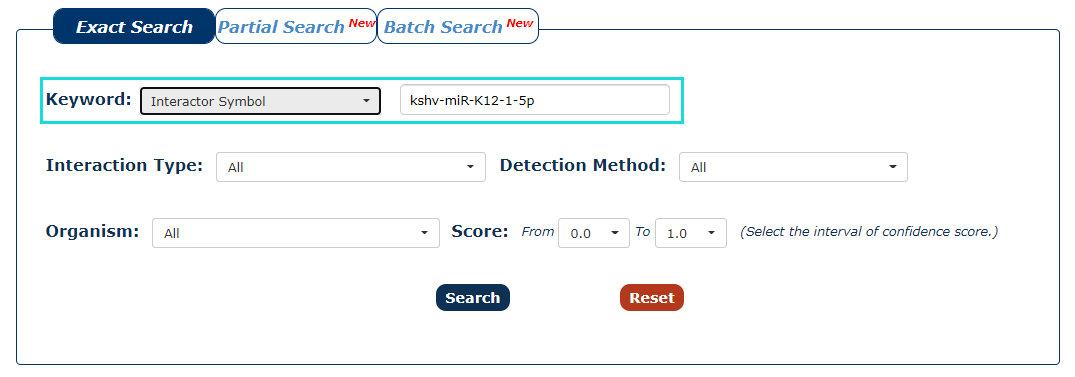
Fig.3-2
3. The next step is to choose the type of interaction. If you want to search for interaction of virus with host, you can choose 'Virus-Host interactions'. In this example, we choose 'All'. Under this condition, we can get miRNA('kshv-miR-K12-1-5p') associated four types of interactions.
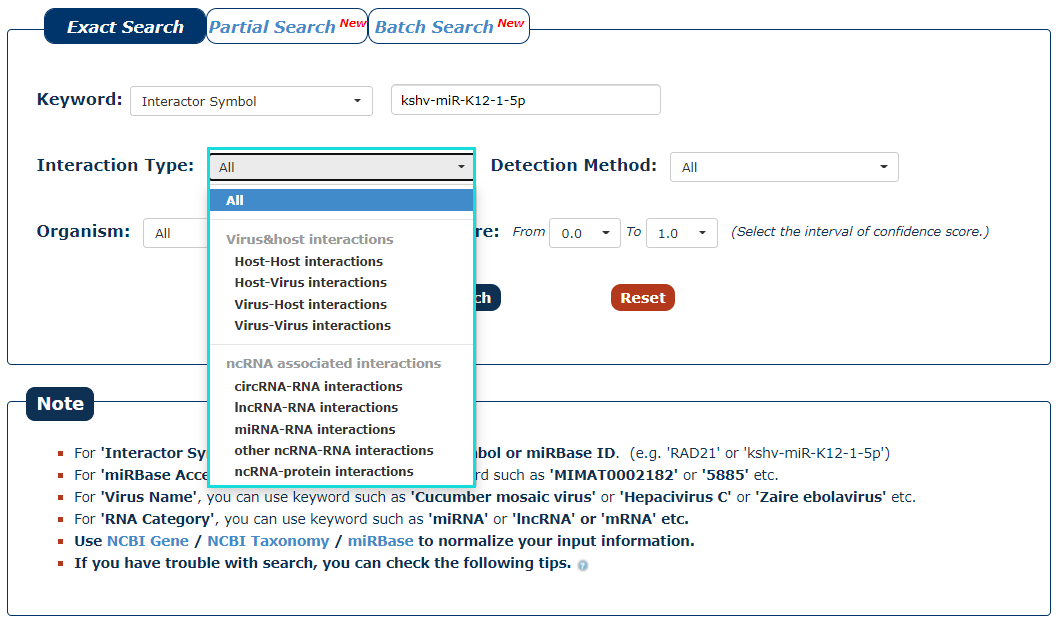
Fig.3-3
4. You can also choose the type of method that detects interaction as the filter. In this example, we want query the interaction detected by strong experimental evidence, so we choose 'Strong Experimental Evidence'.
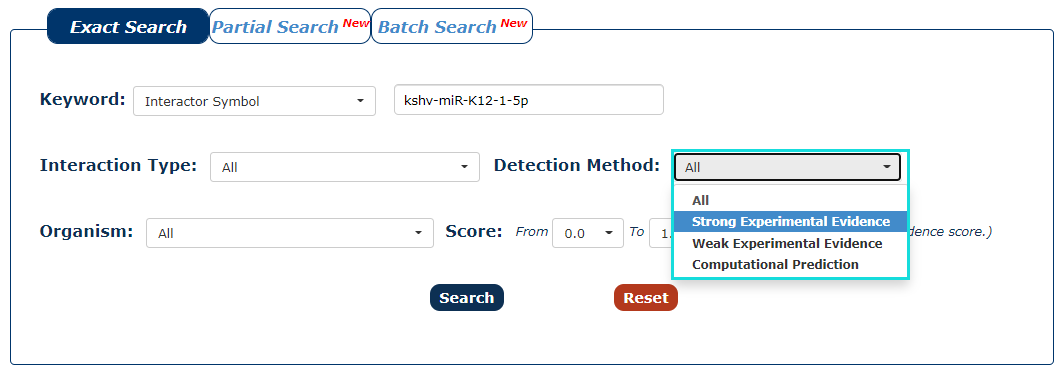
Fig.3-4
5. Then select the organism for the keyword you entered. In this example, we choose 'Homo sapiens' as the organism of the keyword 'kshv-miR-K12-1-5p'.
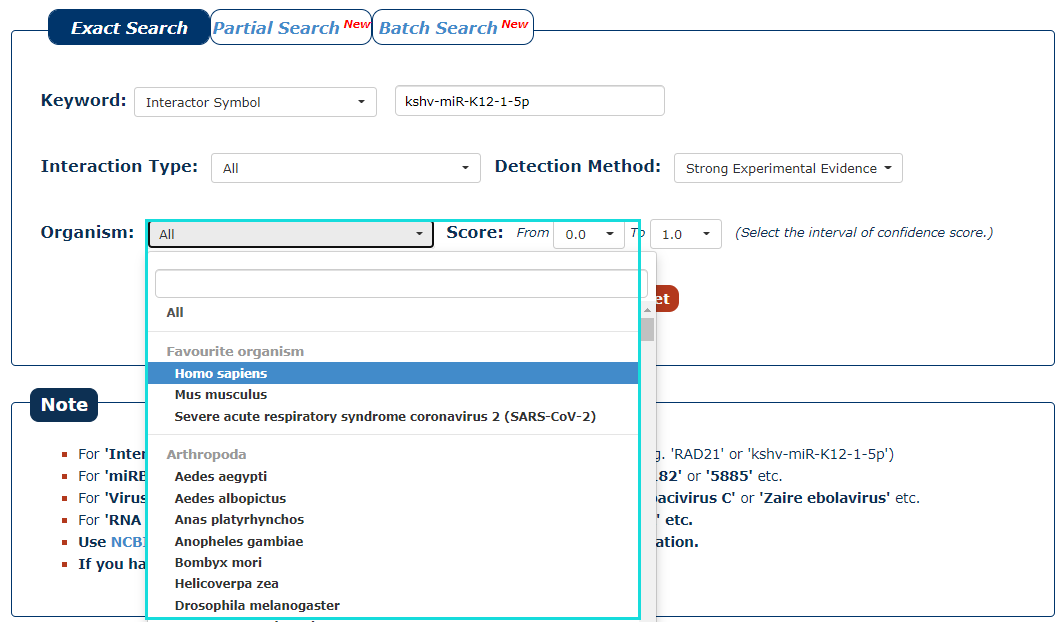
Fig.3-5
6. We provide a score for each interaction. The greater the value, the higher the credibility. To filter low-confidence interactions, in this example, we choose the 0.5 as the minimum score and 1.0 as the maximum score.
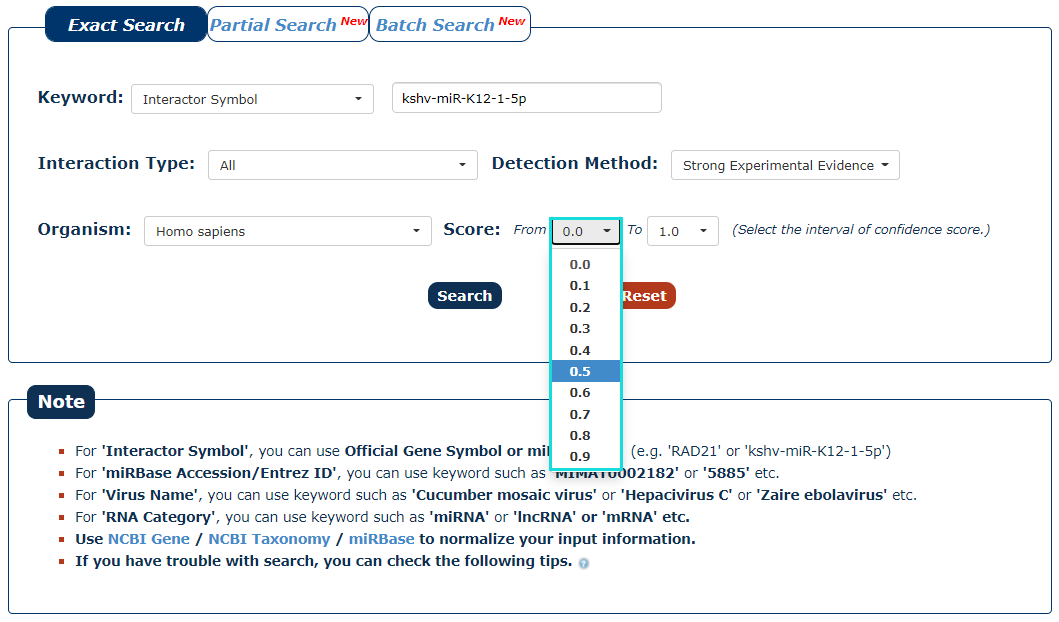
Fig.3-6
7. With all the filters above, we can click 'Search' to query the result.
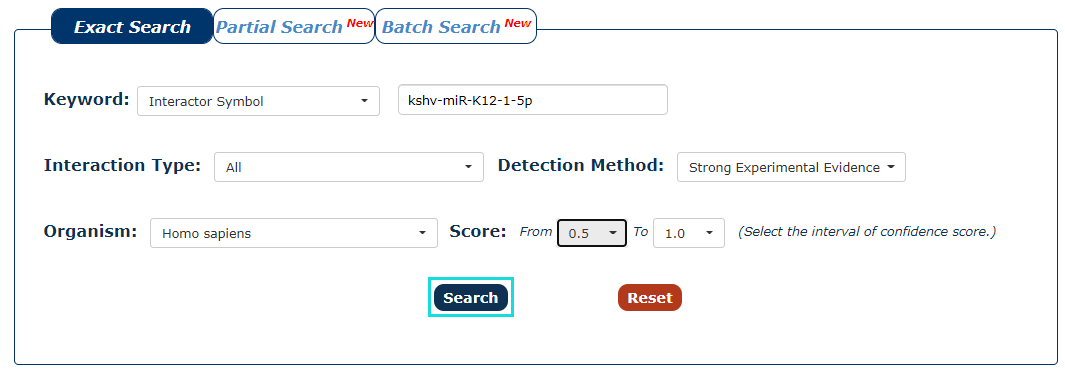
Fig.3-7
8. After several seconds, the result will occur. All the interactions are represented in th table format, and your filters and the total numbers of interactions are in the head of the web page.
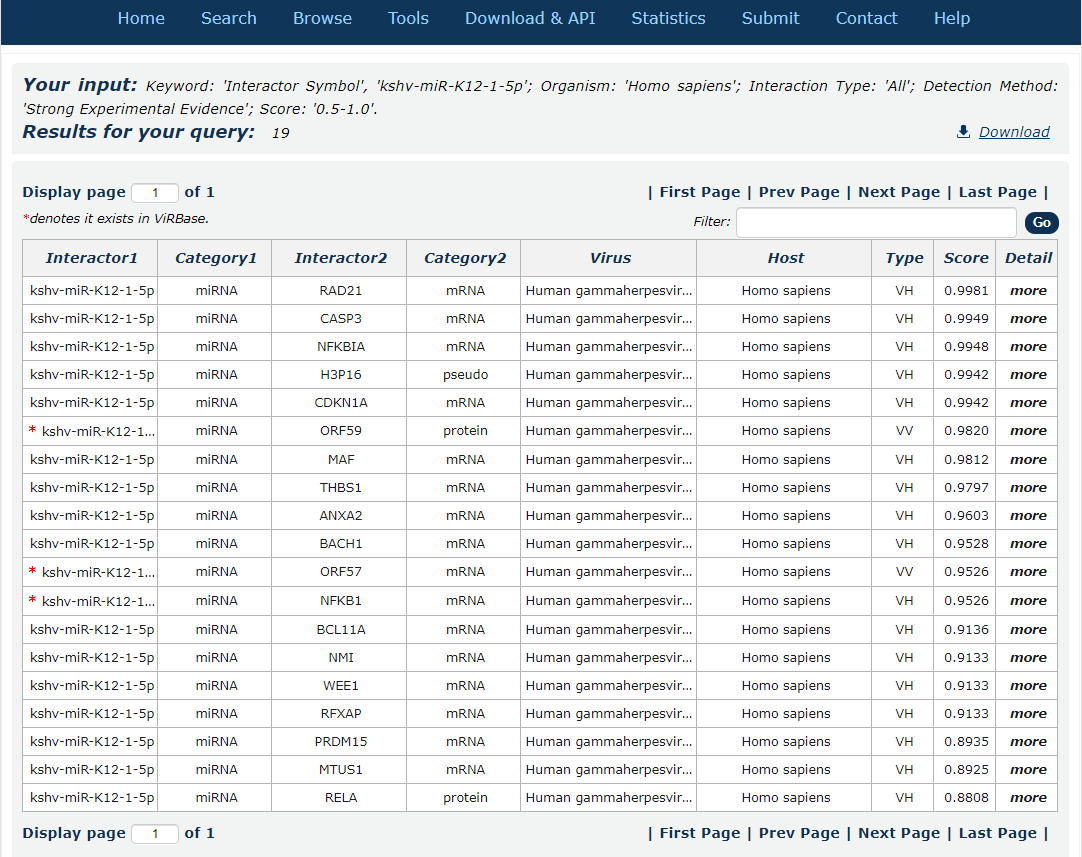
Fig.3-8
In the result page, all entries are listed with basic information including ncRNA symbols,ncRNA categories, virus and host organism, interaction types and score.
Fig.4-1:
1. Your current input conditions.
2. Total sum of results.
3. Download the results.
4. Click to turn the page.
5. Filter the results.
6. Click any interactor as a keyword to search in ViRBase database.
7. Click to link to detail page.
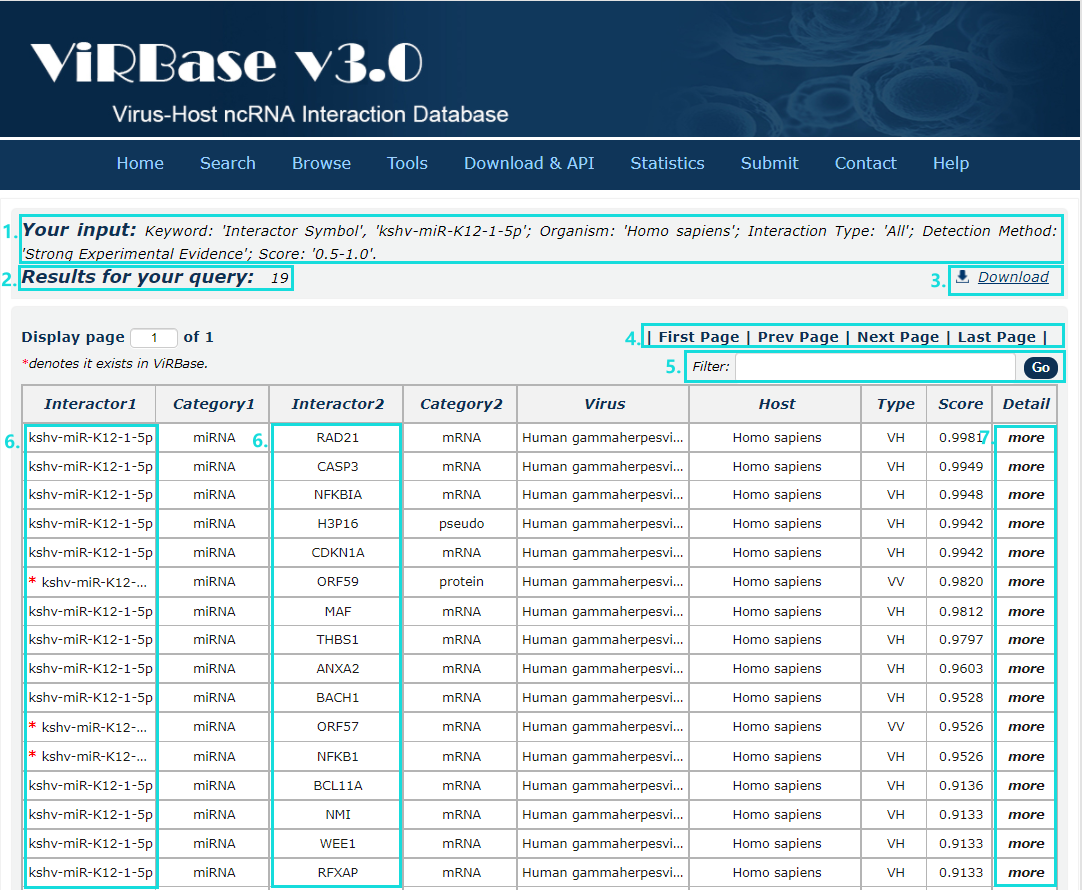
Fig.4-1 Result page
First, you can get general information including ViRBase ID, confidence score, interaction type and predicted binding sites in the detail page.
Second, you can also get the basic information, drug information, the interaction network, evidence support and references of each entry.
Thrid, the association of RNA editing, localization, modification and ncRNA SNP also been provided.
Fig.5-1:
For ncRNA-RNA interactions, users can choose any union of two transcript accessions and click to see results displayed either by miRanda or RIsearch.
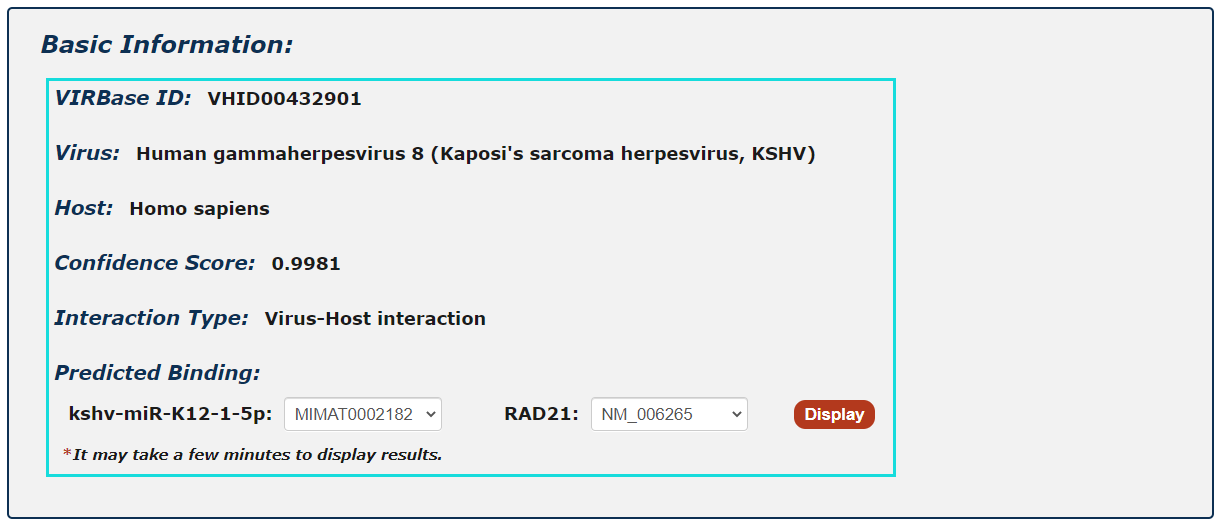
Fig.5-1 Detail page of RNA-RNA interaction
Fig.5-2:
1. For ncRNA-Protein interactions, users can choose any protein sequence accession and click to see results displayed by PRIdictor.
2. Users can also get ncRNA-binding sites in proteins documented in RBPDB, RsiteDB and PDB.
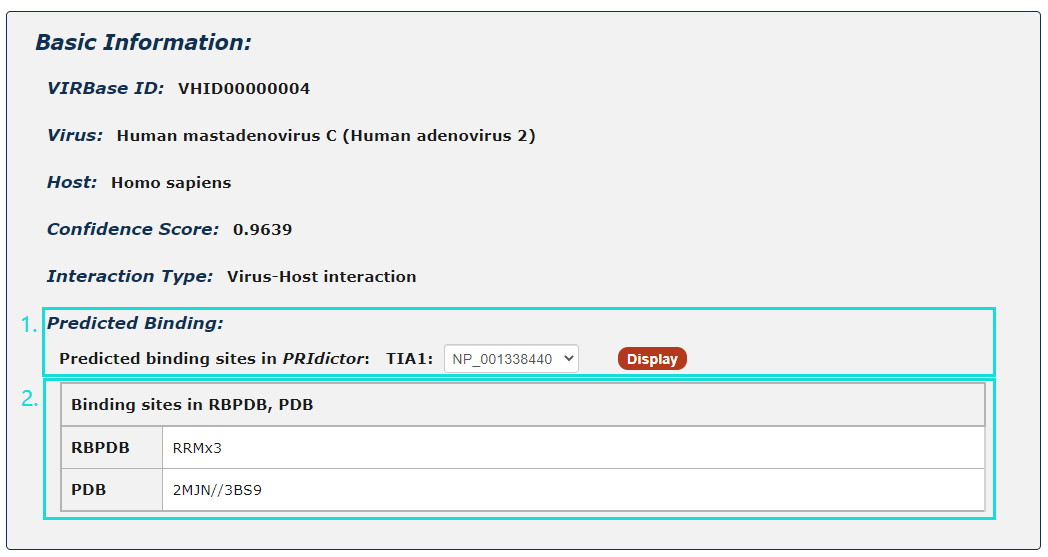
Fig.5-2 Detail page of ncRNA-Protein interaction
Fig.5-3:
1. Click any interactor as a keyword to search in database.
2. Click Entrez ID/miRBase Accession to see its basic description in NCBI Gene/miRbase database.
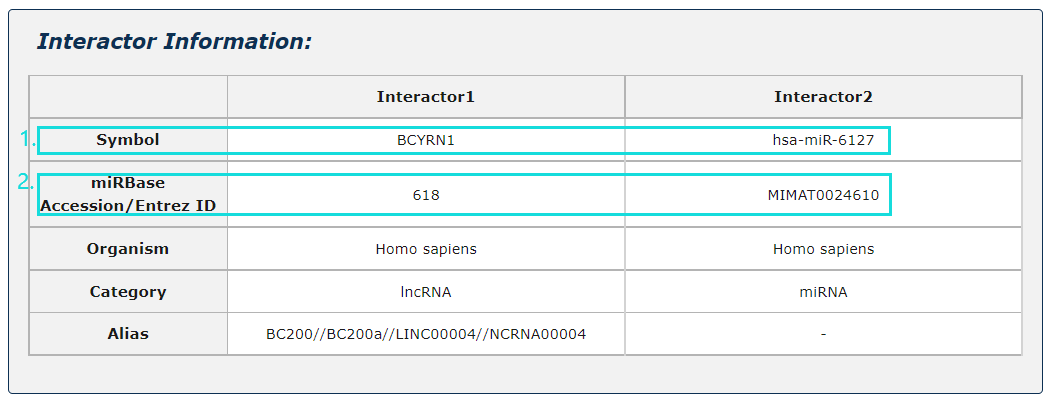
Fig.5-3 Detail page of basic information
Fig.5-4:
1. ncRNA SNP information from two source is provided.
2. For SNP site, provide SNP ID.
3. For SNP site, provide SNP position.
4. For SNP site, provide the variety of base site.
5. For SNP site, provide Pubmed ID.
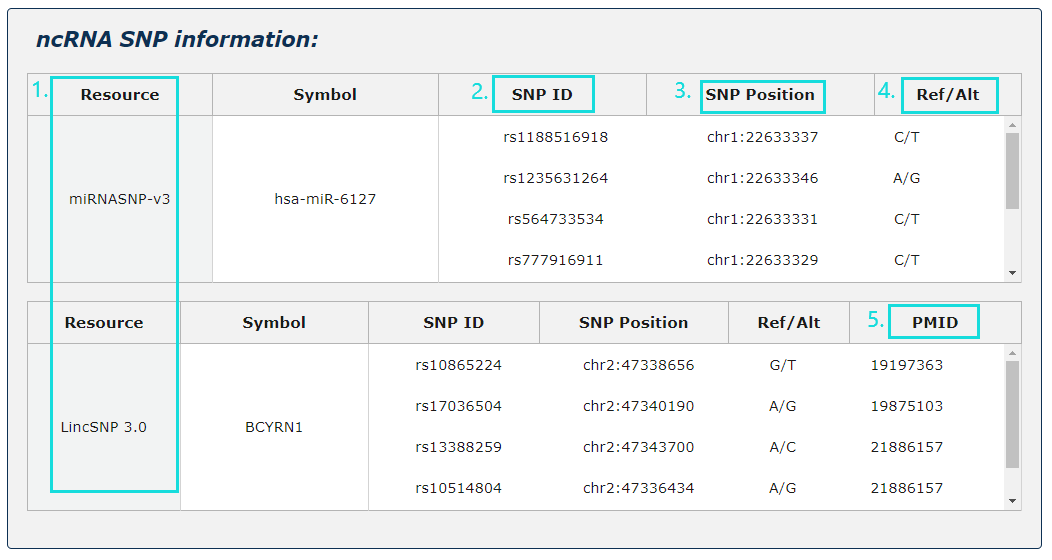
Fig.5-4 Detail page of ncRNA SNP information
Fig.5-5:
1. RNA editing information from three source is provided.
2. For Lncediting, provide editing position.
3. For RADAR, provide editing position, change and genetic region.
4. For DARNED, provide editing position, change, seqReg, exReg and PubMed ID.
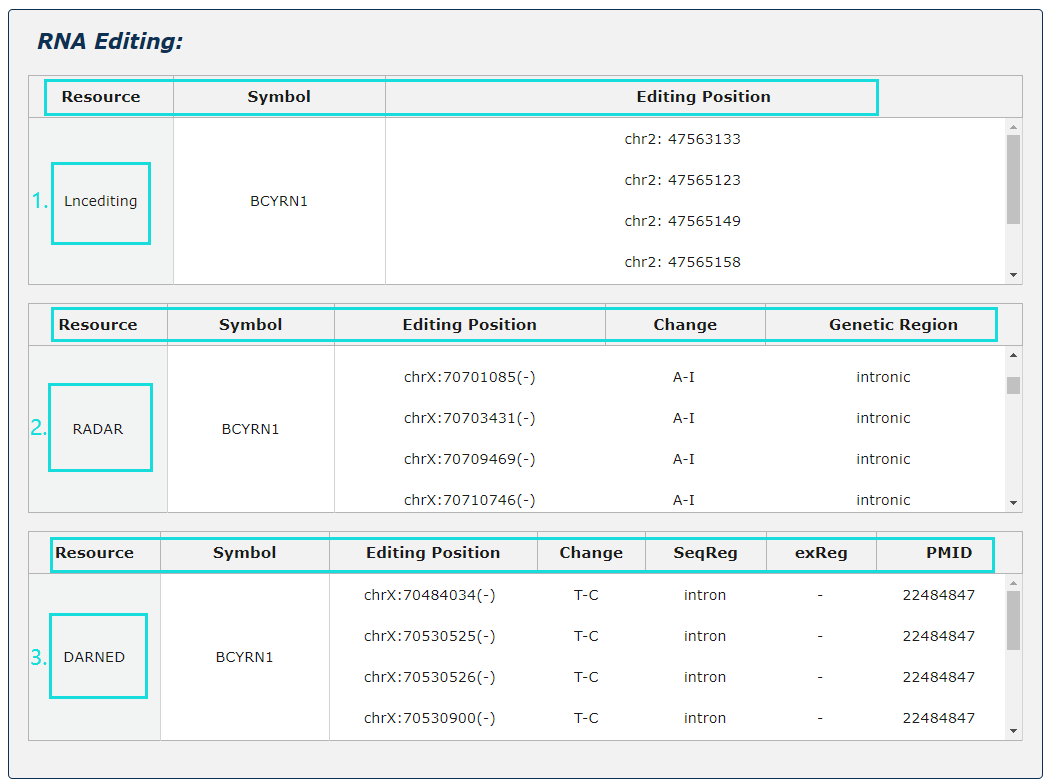
Fig.5-5 Detail page of RNA editing
Fig.5-6:
RNA modification information from RMBase is provided, include modification positions, modification types and genomic contexts for each RNA symbol.
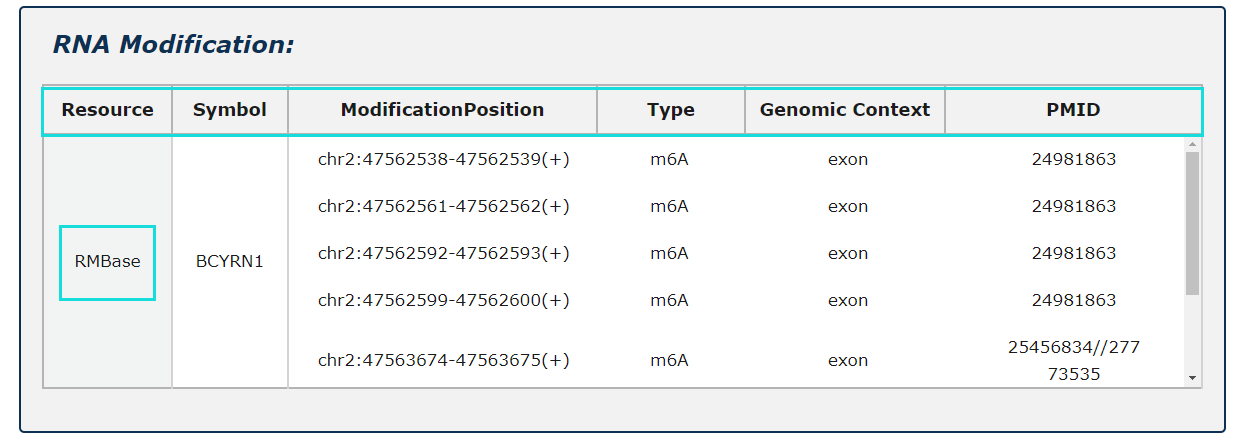
Fig.5-6 Detail page of RNA modification
Fig.5-7:
RNA localization information from RNALocate is provided, include symbol, subcellular localization, tissue or cell line, PMID. Click each subcellular localization can jump to RNALocate database.
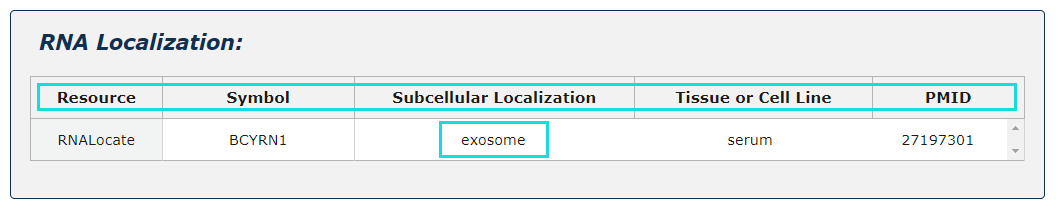
Fig.5-7 Detail page of RNA localization
Fig.5-8:
1. Related drug information from four source is provided.
2. Four functions of RNA-Drug are demonstrated.
3. Click Link and PubChem ID can see more information.
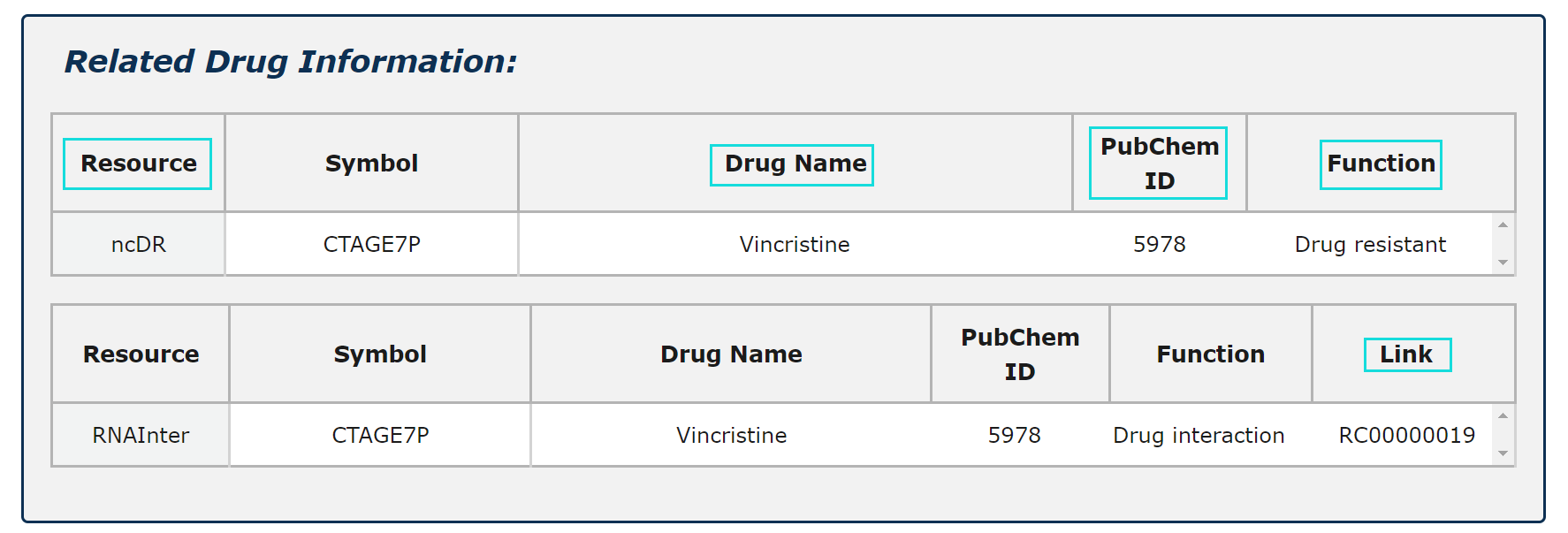
Fig.5-8 Detail page of drug information
Fig.5-9:
1. Provide interaction network for each interactor (show the top 100 interactions ranked by confidence score in our database).
2. Click each iron can drop this interactor category during generate network plot.
3. Click each edge will redirect to corresponding detail page of interaction data.
4. The interaction network plot can be download.
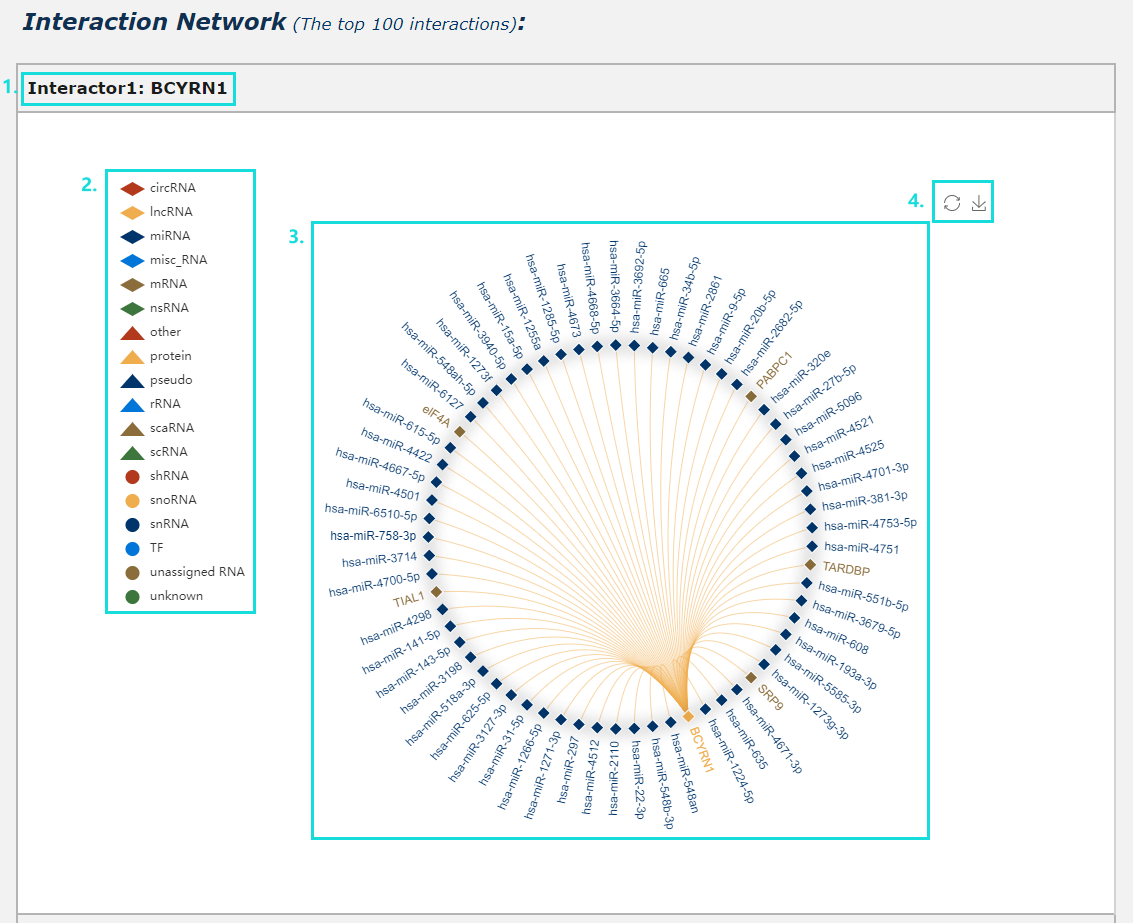
Fig.5-9 Detail page of RNA secondary structure
Fig.5-10:
1. Evidence support including four parts: strong evidence, weak evidence, prediction evidence and support database.
2. The source of database, tissue or cell line and description of each entry.
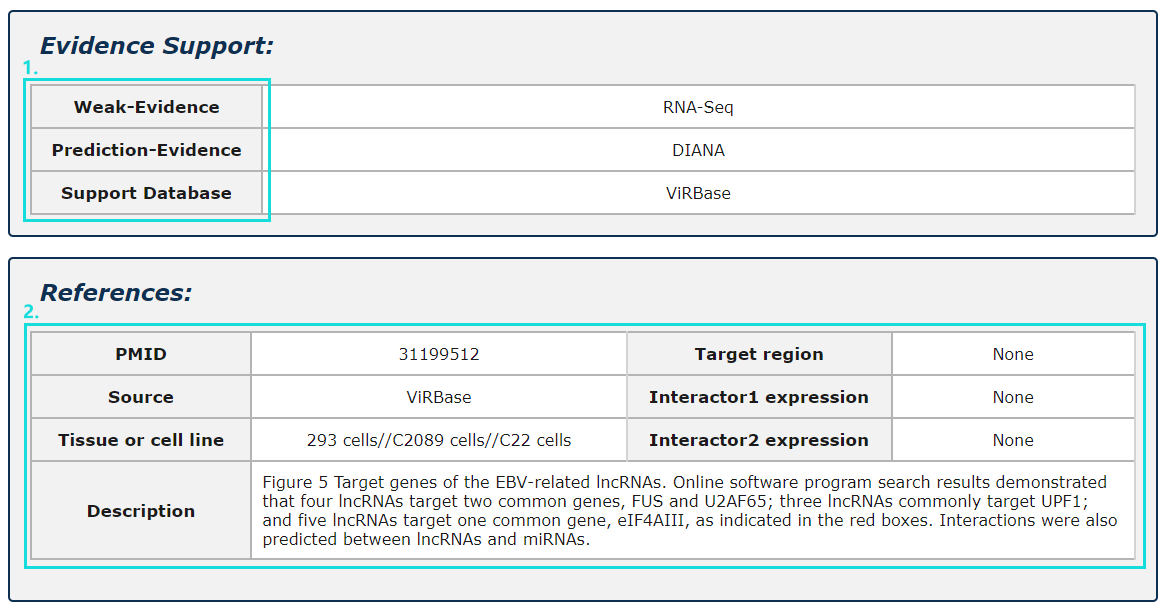
Fig.5-10 Detail page of evidence support
In the browse page, you can click each node to see results.
1. 'Interaction type' indicates the category of all interactors.
2. 'Detection methods' display all entries as long as the current selected method is involved.
3. 'Organism' display all entries as long as one molecule's organism matches the condition.
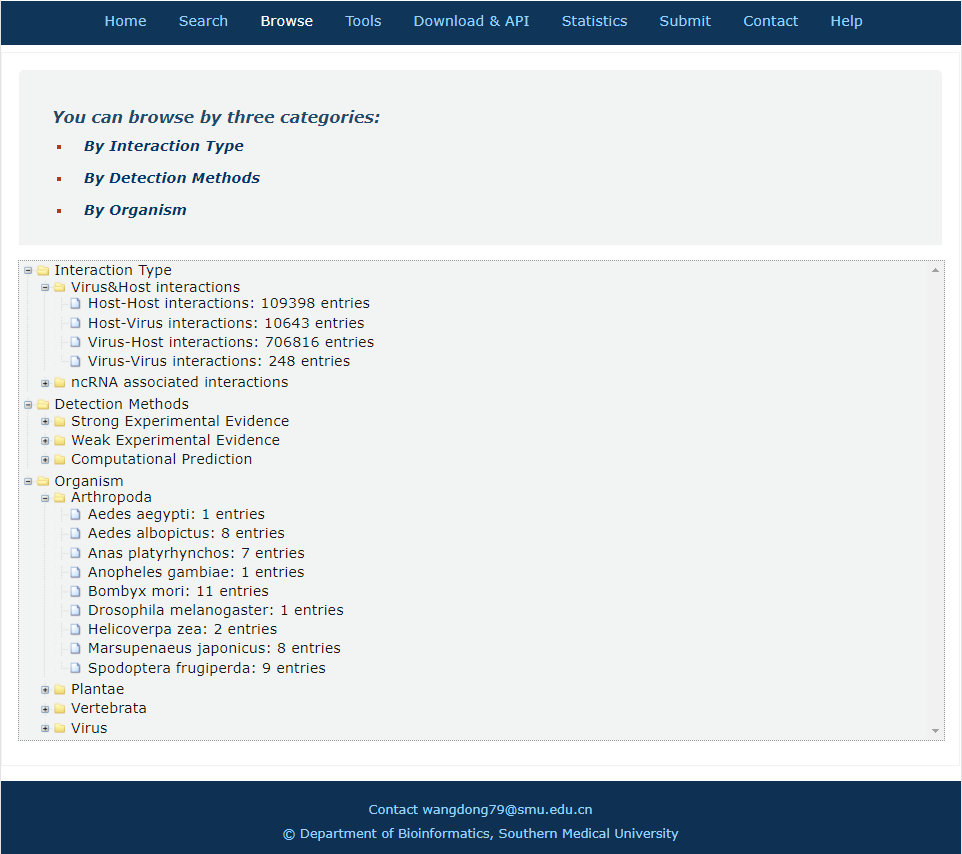
Fig.6-1 Browse page
Two prediction tools are provided in our database, including RNA-RNA and ncRNA-protein.
Fig.7-1:
A program for the fast and accurate prediction of interactions between two RNA molecules is provided.
1. Input RNA sequences or upload files with FASTA format.
2. Select and check the parameters.
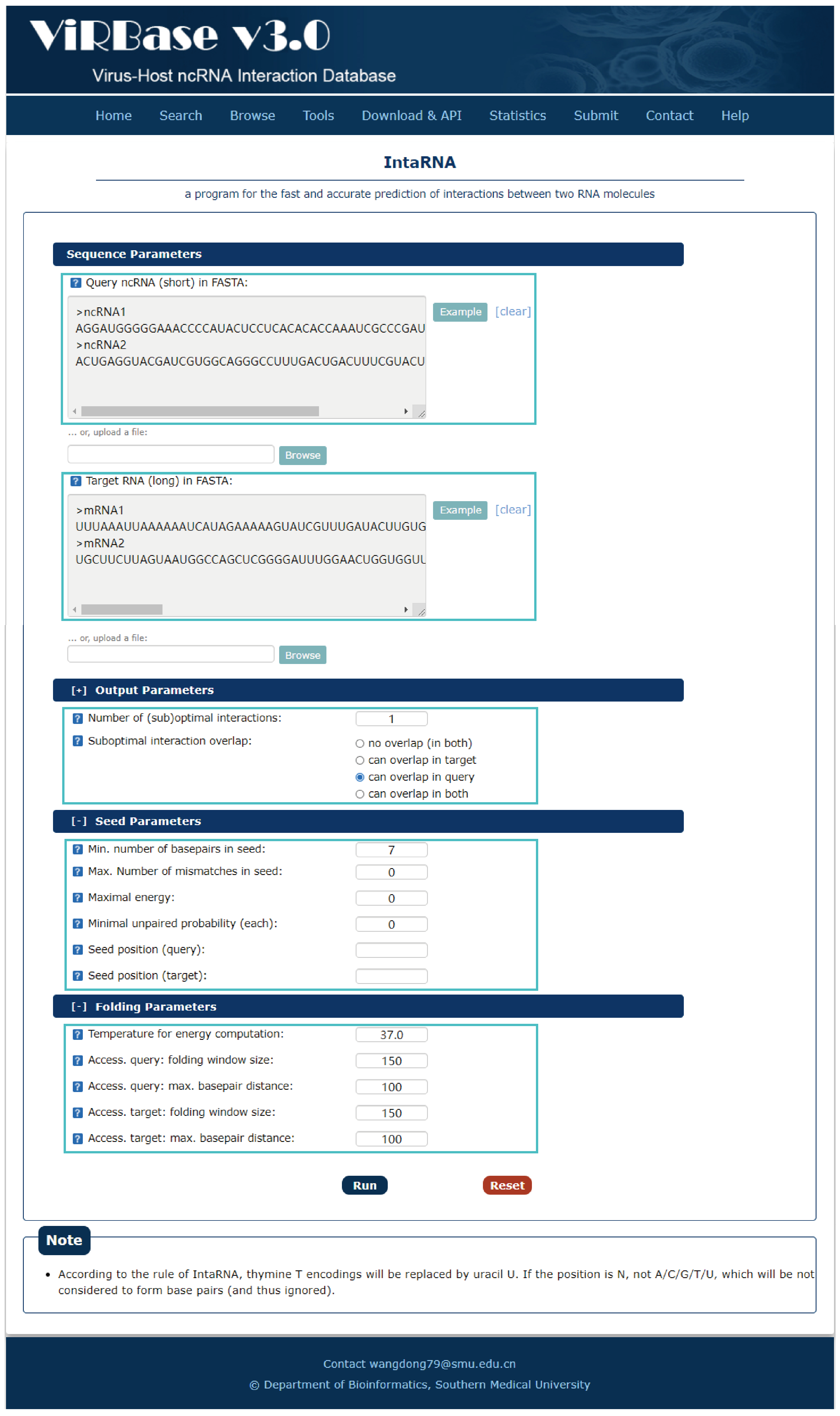
Fig.7-1 Tool of IntaRNA
Fig.7-2:
The search results include target, query, position and energy.
1. The results can be download.
2. Click each result (filling with yellow) can see the interaction position in detail.
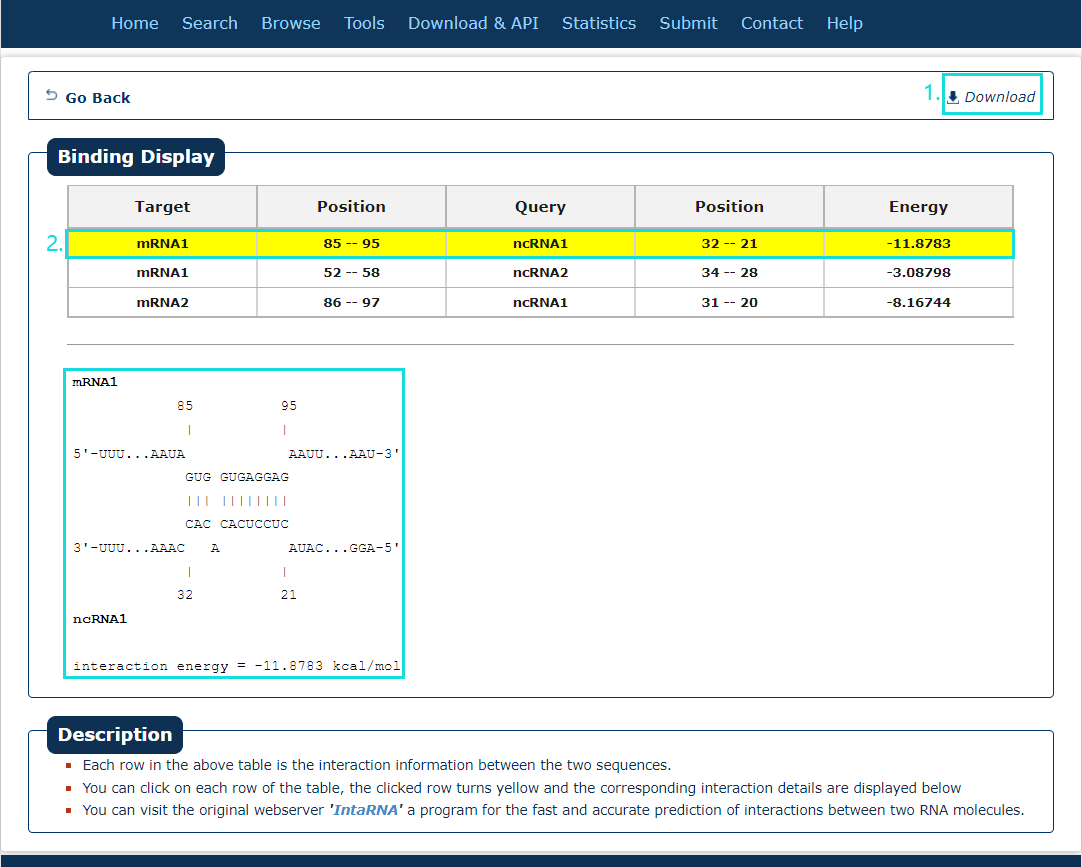
Fig.7-2 Result of IntaRNA
Fig.7-3:
A tool for Protein-RNA interaction predict is provided.
1. Input RNA and(or) protein sequence(s).
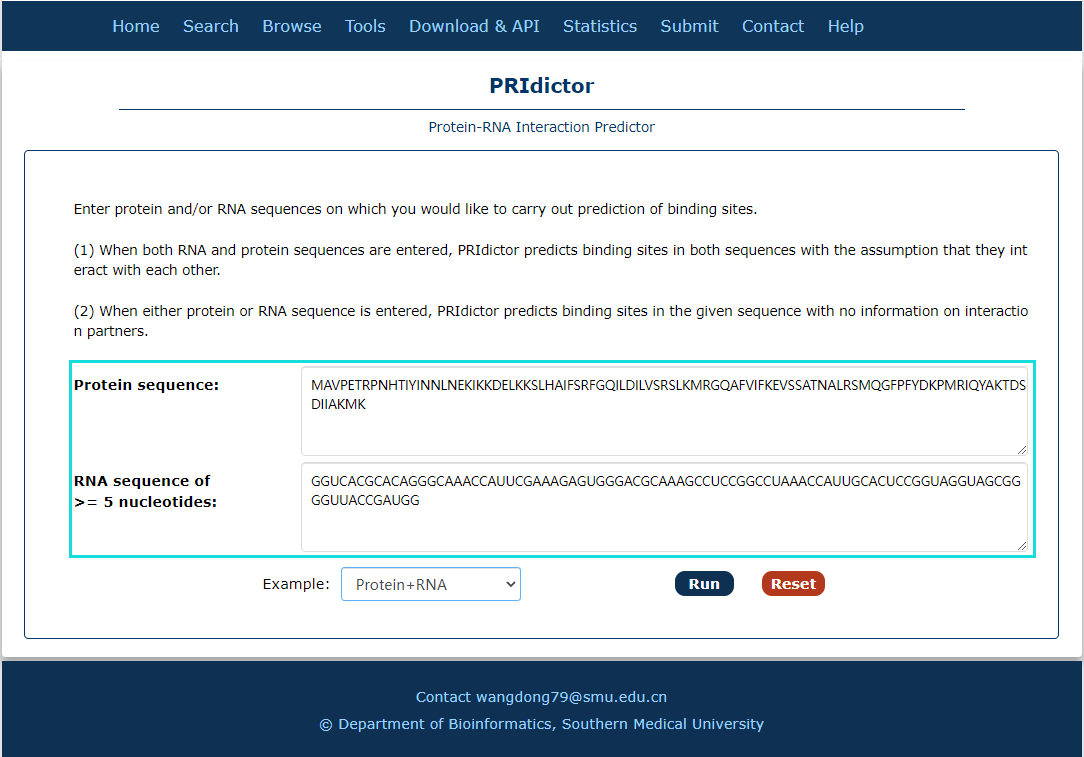
Fig.7-3 Tool of PRIdictor
Fig.7-4:
The search results include sequence, confidence and prediction sites.
1. The results can be download.
2. Every site correspond a score, click column can see in histogram.
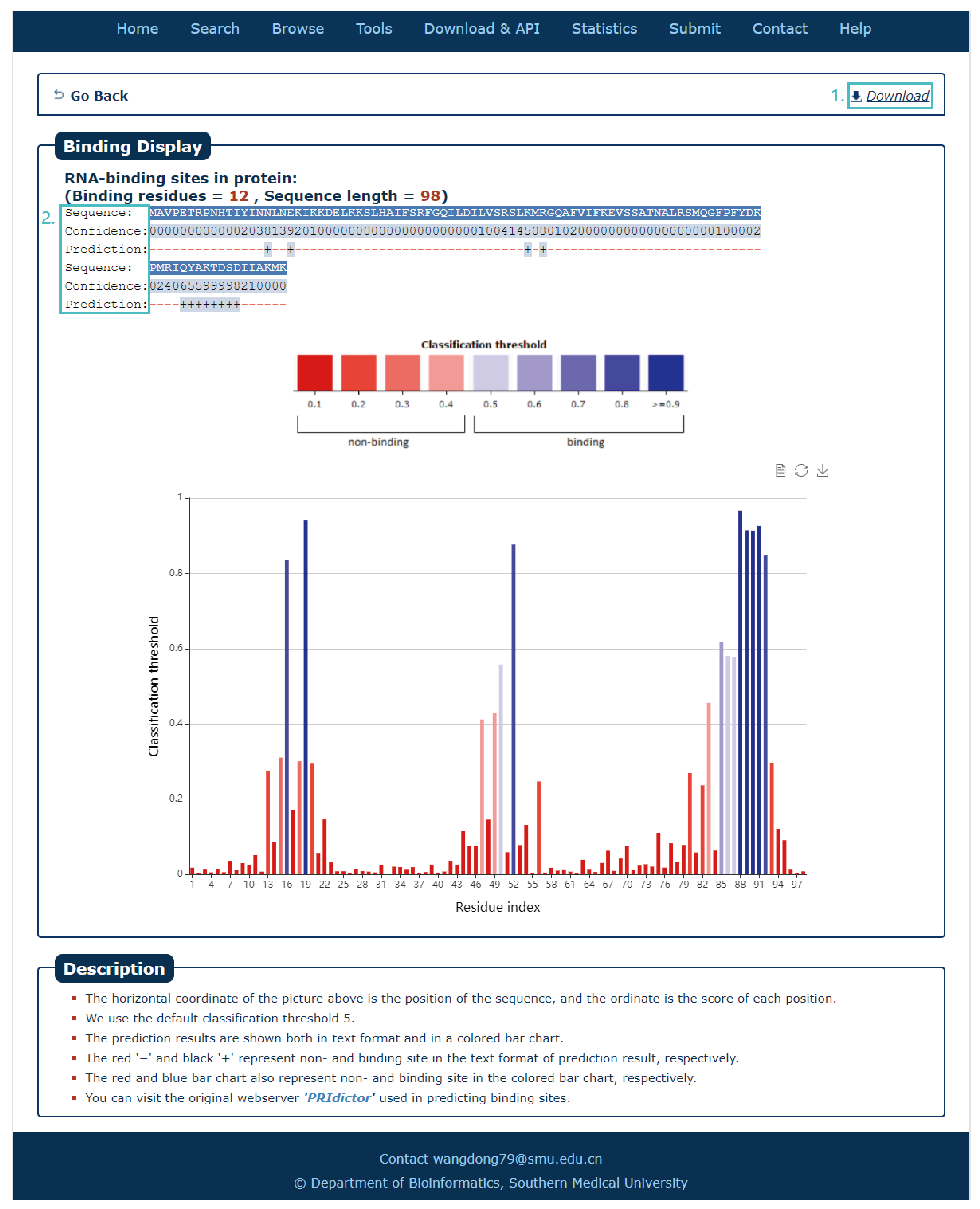
Fig.7-4 Result of PRIdictor
In ViRBase v3.0, the virus-host ncRNA associated interactions are collected from different types of resources under one common framework, including experimental and prediction evidence. In principle, we assume that:
1. Experimental evidence should contribute more important to the confidence score than prediction evidence;
2. Strong experimental evidence should provide more reliable evidence than weak experimental evidence;
3. virus-host ncRNA associated interactions supported by more evidence should be given significantly higher confidence scores than those supported by fewer evidence.
Similar to RNAInter database, according to the evidence types and number of evidence resources, we calculate the confidence score (S) for each ncRNA-associated interaction as follows:
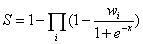
where i is the evidence type(ss: strong experimental evidence, sw: weak experimental evidence, sp: computational prediction method), x is the number of evidence resources, we set weight factor Ws, Ww and Wp to 1, 0.65, and 0.25, respectively(if x=0, we set weight factor Wi to 0).
Fig.8-1:
The score threshold.
According to the distribution of scores, if an interaction entry only has predicted evidences, the score is less than 0.4; if an entry has at least one weak evidence but no strong evidence, the score mainly ranges from 0.4 to 0.7; if an entry has at least one strong evidence, the score ranges from 0.7 to 1.0, the 0.4 and 0.7 could be used as thresholds for choosing the interactions.
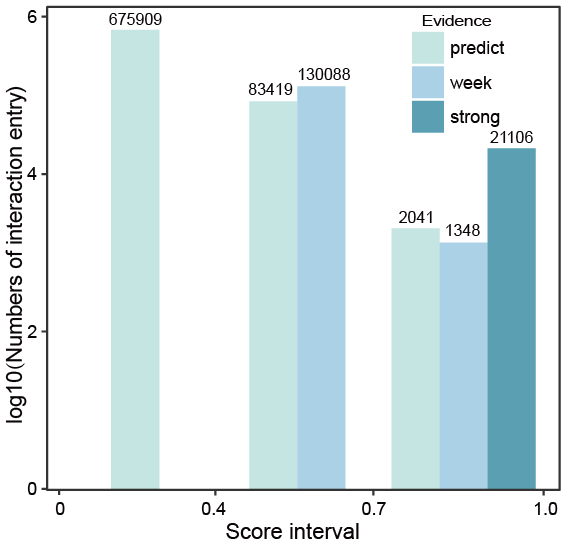
Fig.8-1 The score threshold
Integration of source databases which use different interactors naming conventions is challenging. To ensure maximal connectivity of data, we transform each interactor name found in the input sources to the appropriate naming convention.
1. For miRNA, we use miRBase ID and miRBase Accession.
2. For compound, we use NCBI PubChem Compound symbol.
3. For others, we use official Gene Symbol and Entrez ID.
4. For organism, we normalized organism names according to NCBI Taxonomy Database.