- Source
- Tutorial
- Parameter
- Error Report
Our Relevant Work
- miRDeathDB (Xu J, Li YH et al.,Cell Death Differ. 2012 Sep)
- MNDR (Yuting Wang, Dong Wang, et.al.,Cell Death Disease. 2013.)
- RAID (Xiaomeng Zhang, Dong Wang, et.al.,RNA. 2014.)
ncRNA Related Databases
- human microRNA disease database (HMDD) (Lu M, Zhang Q et al.,PLoS ONE.2008 Oct)
- lncrnadb (Amaral PP et al.,Nucleic Acids Res 39: D146-151.)
- miRTarBase (Hsu SD et al.,Nucleic Acids Res. 2011 Jan)
- miR2Disease (Jiang Q, Wang Y et al.,Nucleic Acids Res.2009 Jan:D98-104)
- miRNAMap 2.0 (Hsu SD, Chu CH et al.,Nucleic Acids Res.2008 Jan:D165-9)
- miRbase (Kozomara A et al.,Nucleic Acids Res. 2011 D152-D157)
- NPInter (Wu T et al.,Nucleic Acids Res. 2006 Jan:D150-2)
- ncRNA Expression Database (NRED) (Dinger ME, Pang KC et al.,Nucleic Acids Res.2009 Jan)
- NONCODE (Liu C, Bai B et al.,Nucleic Acids Res.2005 Jan:D112-5)
- PRD (Fujimori S et al.,Bioinformation. 2012)
- RNAdb (Bateman A, Agrawal S et al.,RNA. 2011 Nov;17(11):1941-6.)
- Subviral RNA Database (Rocheleau L, Pelchat M et al.,BMC Microbiol. 2006 Mar)
Binding Site Predict Tools
- bindN+ (Wang, L. and Brown, S.J et.al., BMC Syst Biol. 2010 May)
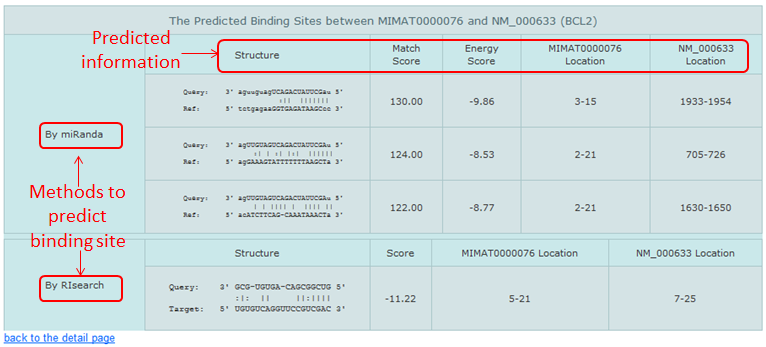
- miranda (John B et al.,PLoS Biol. 2005 Jul)
- RIsearch (Wenzel A et al.,Bioinformatics. 2012 Nov)
- RNAbindR (Terribilini M et al.,Nucleic Acids Research. 2007 Jul)
Other Gene/Protein Sources
Search ncRDeathDB by three paths: Keyword, Death Pathway and Advanced Search
Search Results and Detail Information
Browse
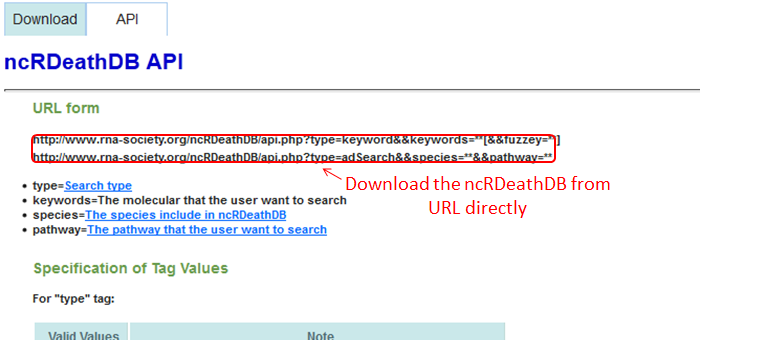
Download & API
Help
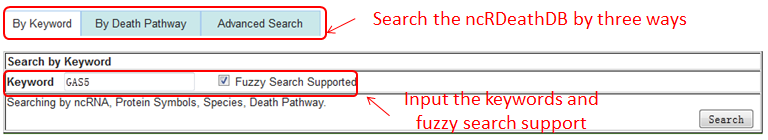

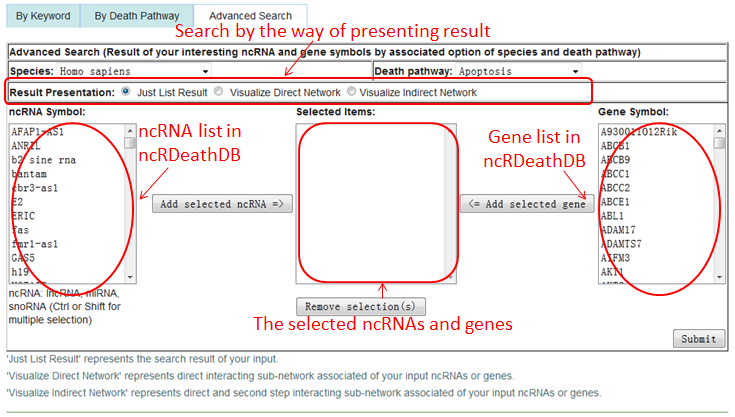
Users can browse and obtain any ncRDeathDB through three paths.
Path 1 (By Keyword): browse the ncRNA-associated cell death entries by inputting the keywords(any ncRNA, Gene Symbol,Death Pathway,Species) and fuzzy search supported.
Path 2 (By Death Pathway): represent all ncRNA-associated cell death entries by associated option of species and death pathway.
Path 3 (By Advanced Search): represent the accurate search results by associated option of species, death pathway and ncRNA/Gene symbols exist in ncRDeathDB. Multiple selection supported by enter the 'Ctrl' or 'Shift'
In this path, users can choose the result presentation modes (including result and network) by select the "Result Presentation"
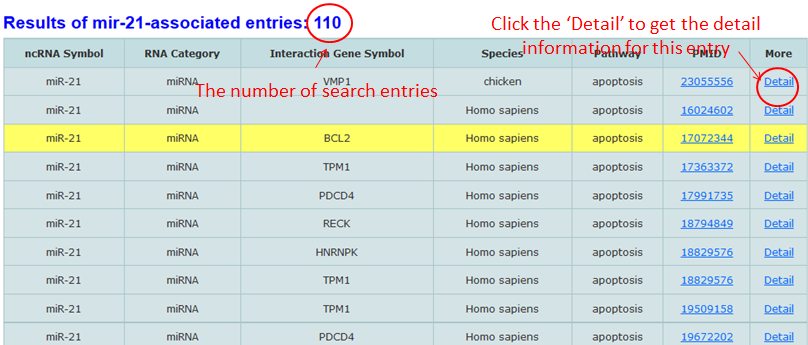

We directly link to snoRNAbase, lncrnadb and NCBI when the ncRNA category is lncRNA or snoRNA.
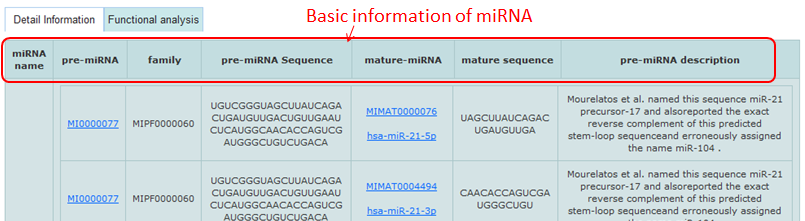
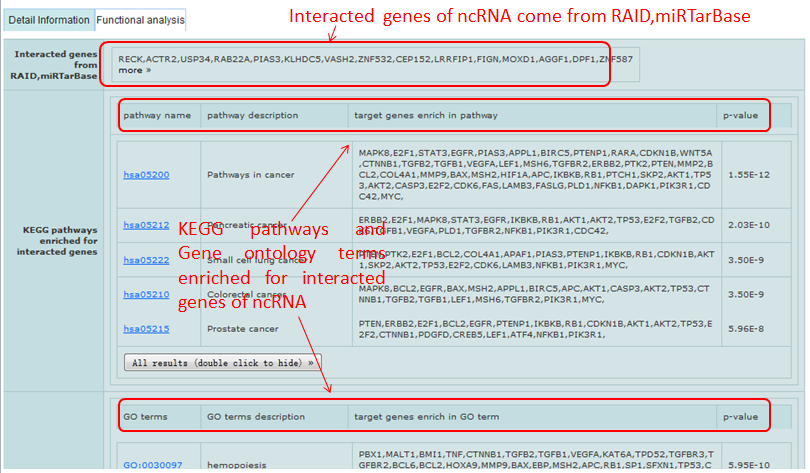
Detail information of miRNAs mainly contains their sequences, family, summary and interacted genes.
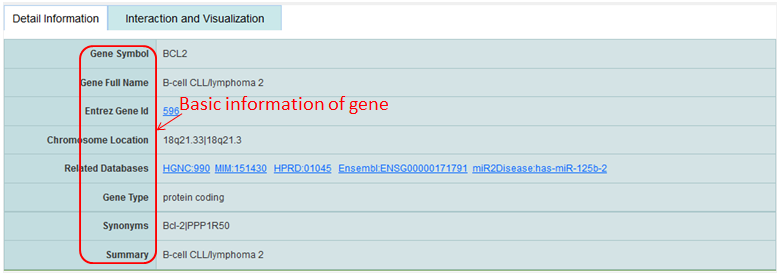
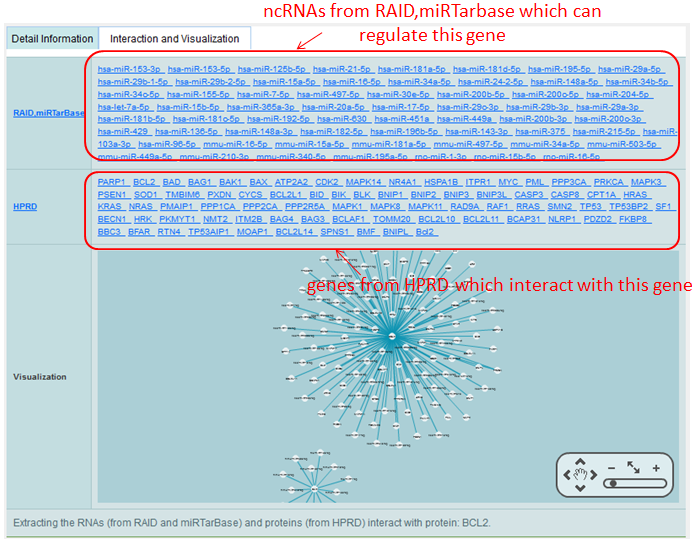
Detail information of genes mainly contains their Synonyms, chromosome location, links to varies of databases and the interacted ncRNAs and genes.
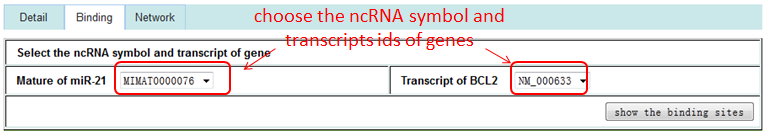
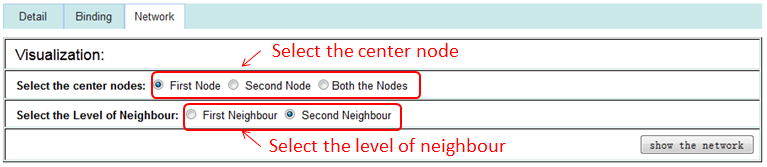
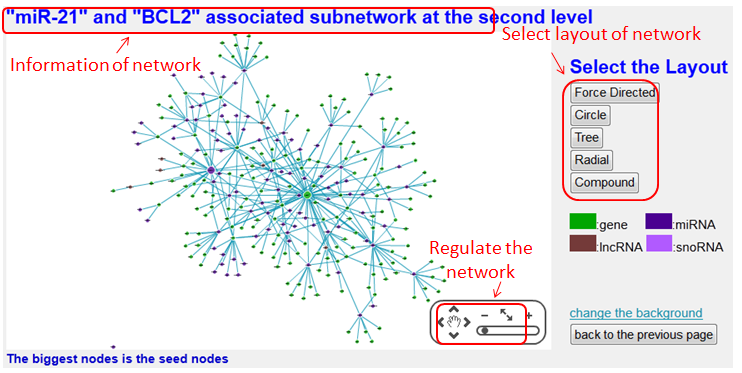
The 'First Node' or 'Second Node' option represents the sub-network of interacting RNA/Protein with the first or second interaction node, the 'Both the Nodes' option represents the sub-network of interacting RNA/Protein with both of interaction nodes.
The 'First Neighbour' represents the sub-network of direct interacting with the center node, the 'Second Neighbour' represents the sub-network of direct or second step interacting with the center node.
Interaction sub-network based on the two nodes of this interaction may help the researchers represents all interacting partners immediately.



bindN+
Web Server: Submit the protein sequences to bindN+ and get the binding locationhttp://bioinfo.ggc.org/bindn+/
Parameter setting
Miranda
Manual Location: The manual and the parameter of Mirandahttp://cbio.mskcc.org/microrna_data/manual.html
Parameter setting